Abstract
There are no disease-modifying treatments for adult human neurodegenerative diseases. Here we test RNA-targeted therapies1 in two mouse models of spinocerebellar ataxia type 2 (SCA2), an autosomal dominant polyglutamine disease2. Both models recreate the progressive adult-onset dysfunction and degeneration of a neuronal network that are seen in patients, including decreased firing frequency of cerebellar Purkinje cells and a decline in motor function3,4. We developed a potential therapy directed at the ATXN2 gene by screening 152 antisense oligonucleotides (ASOs). The most promising oligonucleotide, ASO7, downregulated ATXN2 mRNA and protein, which resulted in delayed onset of the SCA2 phenotype. After delivery by intracerebroventricular injection to ATXN2-Q127 mice, ASO7 localized to Purkinje cells, reduced cerebellar ATXN2 expression below 75% for more than 10 weeks without microglial activation, and reduced the levels of cerebellar ATXN2. Treatment of symptomatic mice with ASO7 improved motor function compared to saline-treated mice. ASO7 had a similar effect in the BAC-Q72 SCA2 mouse model, and in both mouse models it normalized protein levels of several SCA2-related proteins expressed in Purkinje cells, including Rgs8, Pcp2, Pcp4, Homer3, Cep76 and Fam107b. Notably, the firing frequency of Purkinje cells returned to normal even when treatment was initiated more than 12 weeks after the onset of the motor phenotype in BAC-Q72 mice. These findings support ASOs as a promising approach for treating some human neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rigo, F., Seth, P. P. & Bennett, C. F. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv. Exp. Med. Biol. 825, 303–352 (2014)
Pulst, S. M., Nechiporuk, A. & Starkman, S. Anticipation in spinocerebellar ataxia type 2. Nat. Genet. 5, 8–10 (1993)
Dansithong, W. et al. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLoS Genet. 11, e1005182 (2015)
Hansen, S. T., Meera, P., Otis, T. S. & Pulst, S. M. Changes in Purkinje cell firing and gene expression precede behavioral pathology in a mouse model of SCA2. Hum. Mol. Genet. 22, 271–283 (2013)
Pulst, S. M. Degenerative ataxias, from genes to therapies: The 2015 Cotzias Lecture. Neurology 86, 2284–2290 (2016)
Houlden, H. & Singleton, A. B. The genetics and neuropathology of Parkinson’s disease. Acta Neuropathol. 124, 325–338 (2012)
Scoles, D. R. et al. Repeat associated non-AUG translation (RAN translation) dependent on sequence downstream of the ATXN2 CAG repeat. PLoS One 10, e0128769 (2015)
Huynh, D. P., Figueroa, K., Hoang, N. & Pulst, S. M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 26, 44–50 (2000)
Huynh, D. P., Maalouf, M., Silva, A. J., Schweizer, F. E. & Pulst, S. M. Dissociated fear and spatial learning in mice with deficiency of ataxin-2. PLoS One 4, e6235 (2009)
Saugstad, J. A., Marino, M. J., Folk, J. A., Hepler, J. R. & Conn, P. J. RGS4 inhibits signaling by group I metabotropic glutamate receptors. J. Neurosci. 18, 905–913 (1998)
Abdul-Ghani, M. A., Valiante, T. A., Carlen, P. L. & Pennefather, P. S. Metabotropic glutamate receptors coupled to IP3 production mediate inhibition of IAHP in rat dentate granule neurons. J. Neurophysiol. 76, 2691–2700 (1996)
Meera, P., Pulst, S. M. & Otis, T. S. Cellular and circuit mechanisms underlying spinocerebellar ataxias. J. Physiol. (Lond.) 594, 4653–4660 (2016)
Mizutani, A., Kuroda, Y., Futatsugi, A., Furuichi, T. & Mikoshiba, K. Phosphorylation of Homer3 by calcium/calmodulin-dependent kinase II regulates a coupling state of its target molecules in Purkinje cells. J. Neurosci. 28, 5369–5382 (2008)
Iscru, E. et al. Sensorimotor enhancement in mouse mutants lacking the Purkinje cell-specific Gi/o modulator, Pcp2(L7). Mol. Cell. Neurosci. 40, 62–75 (2009)
Wei, P., Blundon, J. A., Rong, Y., Zakharenko, S. S. & Morgan, J. I. Impaired locomotor learning and altered cerebellar synaptic plasticity in pep-19/PCP4-null mice. Mol. Cell. Biol. 31, 2838–2844 (2011)
Tsang, W. Y. et al. Cep76, a centrosomal protein that specifically restrains centriole reduplication. Dev. Cell 16, 649–660 (2009)
Ingram, M. et al. Cerebellar transcriptome profiles of ATXN1 transgenic mice reveal SCA1 disease progression and protection pathways. Neuron 89, 1194–1207 (2016)
Fittschen, M. et al. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 16, 181–192 (2015)
Lee, K. H. et al. Circuit mechanisms underlying motor memory formation in the cerebellum. Neuron 86, 529–540 (2015)
Lang, E. J. et al. The roles of the olivocerebellar pathway in motor learning and motor control. A consensus paper. Cerebellum 16, 230–252 (2017)
Liu, J. et al. Deranged calcium signaling and neurodegeneration in spinocerebellar ataxia type 2. J. Neurosci. 29, 9148–9162 (2009)
Miller, T. M. et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 12, 435–442 (2013)
Skotte, N. H. et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One 9, e107434 (2014)
Carroll, J. B. et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther. 19, 2178–2185 (2011)
Xia, H. et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 10, 816–820 (2004)
Keiser, M. S., Boudreau, R. L. & Davidson, B. L. Broad therapeutic benefit after RNAi Expression vector delivery to deep cerebellar nuclei: implications for spinocerebellar ataxia type 1 therapy. Mol. Ther. 22, 588–595 (2014)
Rodriguez-Lebron, E., Liu, G., Keiser, M., Behlke, M. A. & Davidson, B. L. Altered Purkinje cell miRNA expression and SCA1 pathogenesis. Neurobiol. Dis. 54, 456–463 (2013)
Becker, L. A. et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature http://dx.doi.org/10.1038/nature22038 (2017)
Swayze, E. E. et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Meera, P., Wallner, M. & Otis, T. S. Molecular basis for the high THIP/gaboxadol sensitivity of extrasynaptic GABAA receptors. J. Neurophysiol. 106, 2057–2064 (2011)
Acknowledgements
We thank P. Jafar-Nejad for his contributions to interpreting results and for reading and editing the manuscript, L. Pflieger for contributing to the production of Supplementary Data. This work was supported by grants R01NS33123, R56NS33123 and R37NS033123 from the National Institutes of Neurological Disorders and Stroke (NINDS) to S.M.P., the Noorda foundation to S.M.P., NINDS grants RC4NS073009 and R21NS081182 to D.R.S. and S.M.P., NINDS grant NS090930 to T.S.O., and a gift from Ionis Pharmaceuticals. S.M.P. received grant support from the Target ALS Foundation.
Author information
Authors and Affiliations
Contributions
D.R.S. conceived and designed the study, performed experiments, conducted all ICV injections, analysed all data and wrote the manuscript. M.S. performed all motor-testing experiments and with D.R.S. contributed to blinding of all mouse trials including ASO treatments, motor testing and electrophysiological evaluations. M.D.S. also conducted all qPCR analyses of mouse tissues. P.M. designed and performed all electrophysiological experiments, analysed and interpreted the resulting data, and prepared figures. S.P. prepared all western blots. W.D. conducted the study of SCA2 patient-derived fibroblasts. K.P.F. was in charge of mouse breeding. G.H. led the ASO in silico design, ASO in vitro screening, advised the in vivo screening approach, and provided ASOs. F.R. and C.F.B. contributed to the in vivo screening approach, design of motor phenotype studies, and interpretation of results. T.S.O. designed and helped interpret the electrophysiological analyses. S.M.P. conceived and designed the study with D.R.S. and contributed SCA2 patient-derived fibroblasts. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.M.P. is a consultant for Progenitor Life Sciences and Ataxion Pharmaceuticals. T.S.O. is an employee of F. Hoffmann-La Roche, Ltd. G.H., F.R. and C.F.B are employed by Ionis Pharmaceuticals, which supplied the ASOs used in the study.
Additional information
Reviewer Information Nature thanks R. L. Juliano, J. Rothstein and T. Siddique for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 In vitro screen for ATXN2 ASOs by qPCR.
A total of 152 ASOs were delivered at 4.5 μM to HepG2 cells by electroporation in two 384-well plates. ATXN2 expression was evaluated by qPCR (n = 3 wells per ASO). Shown is the evaluation of the eight best positive hit ASOs for half maximal inhibitory concentration (IC50) determination. Values are mean ± s.d. of ATXN2 quantity relative to total RNA. Cont., scrambled control ASO.
Extended Data Figure 2 Positive hit ASOs evaluated in vivo.
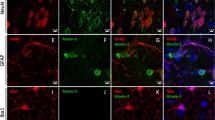
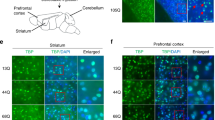
a–c, 250 μg of the indicated ASOs in a total of 7 μl was delivered by ICV injection. After 7 days treatment, the expression of mouse Atxn2 or human ATXN2 and Aif1 was determined by qPCR relative to Actb. a, Wild-type FVB mice. Whereas ASO7 reduced mouse Atxn2 the most by 50%, significant reduction of mouse Atxn2 was not indicated by ANOVA testing for any of ASOs. ASO8 significantly elevated Aif1 expression (P < 0.05). b, BAC-Q72 mice. ATXN2 expression was significantly reduced by ASOs 1, 3 and 7 compared to saline (P < 0.001, 0.01 and 0.01, respectively). Elevations of Aif1 was not observed compared to saline-treated mice. c, ATXN2-Q127 mice. Compared to saline treated mice, ASOs 1, 3, 7 and 8 all significantly lowered ATXN2 expression by 40% or greater (P < 0.001), whereas ASOs 3 and 8 increased Aif1 expression (P < 0.001). Values are mean ± s.d. relative to those determined from normal saline-treated mice. Statistical tests were ANOVA followed by the Bonferroni correction. Technical replication was inserted by employing qPCR with triplicate determinations, and biological replication was made by evaluating multiple mice and/or mouse lines. The number of mice (left-to-right in each chart) was as follows: a, n = 2, 2, 2, 3, 2, 2, 2, 2, 1; b, n = 2, 1, 1, 2, 2, 2, 1, 2; c, n = 2, 3, 1, 1, 1. d, e, ASOs localized to the cerebellar Purkinje cell layer of treated mice. Mice were treated by ICV injection into the right lateral ventricle of the indicated lead ASOs for 7 days in BAC-Q72 mice (ASO3 and ASO7 used at 250 μg) or 10 weeks in ATXN2-Q127 mice (ASO1 used at 200 μg). ASOs were localized in paraffin embedded sections by immunohistochemical peroxidase staining using an anti-ASO antibody. Saline, ATXN2-Q127 mice treated by ICV injection of 7 μl saline for 10 weeks. d, 10× objective. e, 3× digital zoom of a region of the Purkinje cell layer in the corresponding 10× image, indicated by the box. Scale bars, 100 μm (d), 25 μm (e). f–i, Distribution of ASO7 in the cerebellum. f, ASO7 was distributed in Purkinje cell layers throughout the cerebellum (2× objective). g–i, Higher power images for regions indicated in f showed ASO7 localization in Purkinje cells across the cerebellum: g, 10× objective; h, i, 40× objective.
Extended Data Figure 3 Effects of ASO7 on ATXN2 expression in vivo by dose and time.
a–c, Dose response for ASO7 on ATXN2 expression in BAC-Q72 mice treated with ASO7 by ICV injection for 14 days. a, Expression of cerebellar human ATXN2 and mouse Atxn2 determined by qPCR. The 210 μM dose reduced ATXN2 by 63.2% (P < 0.01) and mouse Atxn2 by 44.5% (P < 0.001) compared to the 0 μM dose. The statistical test used was ANOVA followed by the Bonferroni correction. b, Cerebellar Aif1 expression determined by qPCR demonstrated that Aif1 levels were not significantly altered by ASO7 treatment. c, Cerebellar Gfap expression determined by qPCR demonstrated that Gfap levels were not significantly altered by ASO7 treatment. a–c, The replicate number of mice for the saline, 52 μg, 105 μg and 210 μg treatments was 3, 2, 3 and 3, respectively, and the values indicated are mean ± s.d. The experiment was performed once.
Extended Data Figure 4 Weights of mice before and after rotarod testing.
a, Rotarod test of ATXN2-Q127 mice treated with a single ICV dose of 210 μg ASO7. b, Rotarod test of BAC-Q72 mice treated with a single ICV dose of 175 μg ASO7. Weeks of ASO treatments are indicated on the x axes. Mouse weights were unaffected by ASO7 treatment. Significant differences between weights of BAC-Q72 mice compared to wild-type littermates were observed (P < 0.001 for any age group, Student’s t-test). The relevance of mouse weights on motor phenotype testing is discussed in the Supplementary Discussion.
Extended Data Figure 5 ASO7 lowered expression of wild-type and mutant ATXN2 in cultured SCA2 patient-derived fibroblasts.
a, Patient-derived SCA2(CAG35) fibroblasts were transfected with the indicated quantities of ASO7. After 72 h RNA was prepared and total ATXN2 expression was determined by qPCR. Values are mean ± s.d. of 3 technical replicates from single cultures. ATXN2 was reduced by 79% for the 2 μM dose compared to 0 μM (P < 0.001, Student’s t-test). b, To determine ASO7 effect on the expression of non-mutant (CAG22) and mutant (CAG35) ATXN2, RT–PCR reactions were evaluated by agarose gel electrophoresis, with loading controlled for by GAPDH. Both a and b were replicated once yielding nearly the same result.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion and Supplementary Figure 1, the uncropped blots. (PDF 5702 kb)
Supplementary Tables
This file contains Supplementary Tables 1-2. (XLSX 45 kb)
Rights and permissions
About this article
Cite this article
Scoles, D., Meera, P., Schneider, M. et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544, 362–366 (2017). https://doi.org/10.1038/nature22044
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22044
This article is cited by
-
Quantification of Solid Embryonic Cerebellar Graft Volume in a Degenerative Ataxia Model
The Cerebellum (2024)
-
Single-cell epigenomics and spatiotemporal transcriptomics reveal human cerebellar development
Nature Communications (2023)
-
Mitigating a TDP-43 proteinopathy by targeting ataxin-2 using RNA-targeting CRISPR effector proteins
Nature Communications (2023)
-
TR-FRET-Based Immunoassay to Measure Ataxin-2 as a Target Engagement Marker in Spinocerebellar Ataxia Type 2
Molecular Neurobiology (2023)
-
Contribution of Glial Cells to Polyglutamine Diseases: Observations from Patients and Mouse Models
Neurotherapeutics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.