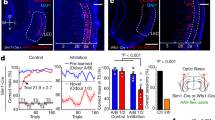
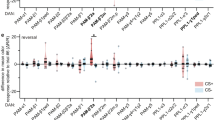
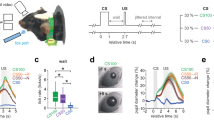
Abstract
Animals constantly assess the reliability of learned information to optimize their behaviour. On retrieval, consolidated long-term memory can be neutralized by extinction if the learned prediction was inaccurate1. Alternatively, retrieved memory can be maintained, following a period of reconsolidation during which it is labile2. Although extinction and reconsolidation provide opportunities to alleviate problematic human memories3,4,5, we lack a detailed mechanistic understanding of memory updating. Here we identify neural operations underpinning the re-evaluation of memory in Drosophila. Reactivation of reward-reinforced olfactory memory can lead to either extinction or reconsolidation, depending on prediction accuracy. Each process recruits activity in specific parts of the mushroom body output network and distinct subsets of reinforcing dopaminergic neurons. Memory extinction requires output neurons with dendrites in the α and α′ lobes of the mushroom body, which drive negatively reinforcing dopaminergic neurons that innervate neighbouring zones. The aversive valence of these new extinction memories neutralizes previously learned odour preference. Memory reconsolidation requires the γ2α′1 mushroom body output neurons. This pathway recruits negatively reinforcing dopaminergic neurons innervating the same compartment and re-engages positively reinforcing dopaminergic neurons to reconsolidate the original reward memory. These data establish that recurrent and hierarchical connectivity between mushroom body output neurons and dopaminergic neurons enables memory re-evaluation driven by reward-prediction error.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dunsmoor, J. E., Niv, Y., Daw, N. & Phelps, E. A. Rethinking extinction. Neuron 88, 47–63 (2015)
Nader, K. Reconsolidation and the dynamic nature of memory. Cold Spring Harb. Perspect. Biol. 7, a021782 (2015)
Kindt, M., Soeter, M. & Vervliet, B. Beyond extinction: erasing human fear responses and preventing the return of fear. Nat. Neurosci. 12, 256–258 (2009)
Schiller, D. et al. Preventing the return of fear in humans using reconsolidation update mechanisms. Nature 463, 49–53 (2010)
Xue, Y. X. et al. A memory retrieval-extinction procedure to prevent drug craving and relapse. Science 336, 241–245 (2012)
Aso, Y. et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3, e04580 (2014)
Owald, D. & Waddell, S. Olfactory learning skews mushroom body output pathways to steer behavioral choice in Drosophila. Curr. Opin. Neurobiol. 35, 178–184 (2015)
Liu, C. et al. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516 (2012)
Burke, C. J. et al. Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–437 (2012)
Owald, D. et al. Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron 86, 417–427 (2015)
Claridge-Chang, A. et al. Writing memories with light-addressable reinforcement circuitry. Cell 139, 405–415 (2009)
Aso, Y. et al. Three dopamine pathways induce aversive odour memories with different stability. PLoS Genet. 8, e1002768 (2012)
Séjourné, J. et al. Mushroom body efferent neurons responsible for aversive olfactory memory retrieval in Drosophila. Nat. Neurosci. 14, 903–910 (2011)
Tempel, B. L ., Bonini, N ., Dawson, D. R . & Quinn, W. G. Reward learning in normal and mutant Drosophila. Proc. Natl Acad. Sci. USA 80, 1482–1486 (1983)
Schwaerzel, M., Heisenberg, M. & Zars, T. Extinction antagonizes olfactory memory at the subcellular level. Neuron 35, 951–960 (2002)
Lagasse, F., Devaud, J. M. & Mery, F. A switch from cycloheximide-resistant consolidated memory to cycloheximide-sensitive reconsolidation and extinction in Drosophila. J. Neurosci. 29, 2225–2230 (2009)
Krashes, M. J. & Waddell, S. Rapid consolidation to a radish and protein synthesis-dependent long-term memory after single-session appetitive olfactory conditioning in Drosophila. J. Neurosci. 28, 3103–3113 (2008)
Kitamoto, T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 47, 81–92 (2001)
Schultz, W ., Dayan, P . & Montague, P. R. A neural substrate of prediction and reward. Science 275, 1593–1599 (1997)
Díaz-Mataix, L., Ruiz Martinez, R. C., Schafe, G. E., LeDoux, J. E. & Doyère, V. Detection of a temporal error triggers reconsolidation of amygdala-dependent memories. Curr. Biol. 23, 467–472 (2013)
Pedreira, M. E., Pérez-Cuesta, L. M. & Maldonado, H. Mismatch between what is expected and what actually occurs triggers memory reconsolidation or extinction. Learn. Mem. 11, 579–585 (2004)
Aso, Y. et al. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577 (2014)
Hige, T., Aso, Y., Modi, M. N., Rubin, G. M. & Turner, G. C. Heterosynaptic plasticity underlies aversive olfactory learning in Drosophila. Neuron 88, 985–998 (2015)
Perisse, E. et al. Aversive learning and appetitive motivation toggle feed-forward inhibition in the Drosophila mushroom body. Neuron 90, 1086–1099 (2016)
Riemensperger, T., Völler, T., Stock, P., Buchner, E. & Fiala, A. Punishment prediction by dopaminergic neurons in Drosophila. Curr. Biol. 15, 1953–1960 (2005)
Sevenster, D., Beckers, T. & Kindt, M. Prediction error demarcates the transition from retrieval, to reconsolidation, to new learning. Learn. Mem. 21, 580–584 (2014)
Merlo, E., Milton, A. L., Goozée, Z. Y., Theobald, D. E. & Everitt, B. J. Reconsolidation and extinction are dissociable and mutually exclusive processes: behavioral and molecular evidence. J. Neurosci. 34, 2422–2431 (2014)
Reichelt, A. C., Exton-McGuinness, M. T. & Lee, J. L. Ventral tegmental dopamine dysregulation prevents appetitive memory destabilization. J. Neurosci. 33, 14205–14210 (2013)
Steinberg, E. E. et al. A causal link between prediction errors, dopamine neurons and learning. Nat. Neurosci. 16, 966–973 (2013)
Chang, C. Y. et al. Brief optogenetic inhibition of dopamine neurons mimics endogenous negative reward prediction errors. Nat. Neurosci. 19, 111–116 (2016)
Friggi-Grelin, F. et al. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 54, 618–627 (2003)
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2, 991–1001 (2012)
Krashes, M. J. et al. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 139, 416–427 (2009)
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013)
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014)
Hoopfer, E. D., Jung, Y., Inagaki, H. K., Rubin, G. M. & Anderson, D. J. P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. eLife 4, e11346 (2015)
Folkers, E ., Drain, P. & Quinn, W. G. Radish, a Drosophila mutant deficient in consolidated memory. Proc. Natl Acad. Sci. USA 90, 8123–8127 (1993)
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. ScanImage: flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13 (2003)
Shang, Y., Claridge-Chang, A., Sjulson, L., Pypaert, M. & Miesenböck, G. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell 128, 601–612 (2007)
Wu, J. S. & Luo, L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protocols 1, 2110–2115 (2006)
Hirano, Y. et al. Shifting transcriptional machinery is required for long-term memory maintenance and modification in Drosophila mushroom bodies. Nat. Commun. 7, 13471 (2016)
Acknowledgements
We thank G. Rubin, FlyLight, Bloomington Stock Center and D. J. Anderson for flies. We are grateful to members of the Waddell group and G. Wright for discussion and comments on the manuscript. J.F. was supported by the Deutsche Forschungsgemeinschaft (FE 1563/1-1), S.L. an EMBO Long-Term Fellowship and O.B the Medical Research Council, University College War Memorial Studentship and a Goodger and Schorstein Scholarship. S.W. is funded by a Wellcome Trust Principal Research Fellowship in the Basic Biomedical Sciences, Gatsby Charitable Foundation, Oxford Martin School and Bettencourt–Schueller Foundation.
Author information
Authors and Affiliations
Contributions
J.F. and S.W. conceived the project and designed all experiments. S.L. performed initial extinction experiments. J.F. performed and analysed all behavioural experiments with help from P.C. O.B. performed imaging experiments assisted by J.F. Live imaging data were analysed by O.B. and P.C. The manuscript was written by S.W. and J.F. with comments from P.C. and O.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Extinction and reconsolidation of reward memory requires distinct subsets of dopaminergic neurons that are driven by recurrent and hierarchical connections within the mushroom body output network (related to Figs 1–4).
a, Aversively reinforcing DANs in the paired posterior lateral 1 (PPL1) cluster innervate discrete regions of the vertical mushroom body lobe whereas individual rewarding DANs in the protocerebral anterior medial (PAM) cluster innervate unique zones on the horizontal lobe. b, Each zone innervated by a particular DAN houses the dendritic field of a corresponding MBON. Aversive DANs overlap with the dendrites of MBONs directing behavioural approach whereas rewarding DANs overlay the dendrites of MBONs driving avoidance. c, The presynaptic fields of many MBONs overlap with the dendrites of DANs that innervate the same mushroom body zones, suggesting the presence of local recurrent feedback loops. d, The weight of behavioural drive to approach- and avoidance-directing MBONs is balanced in naive flies (orange and blue circles of equal size). e, Sugar conditioning engages rewarding DANs that innervate the tips of the horizontal lobes of the mushroom body and that drive depression of synaptic connections between odour-activated mushroom body Kenyon cells and MBONs. f, Following reward conditioning the CS+ drive to avoidance-directing MBONs is reduced (smaller orange circle) thereby favouring activation of odour-driven behavioural approach pathways. g, The reward-learning-induced skew in the MBON network is expressed when flies re-encounter the CS+ odour. Preferential CS+ drive of approach-directing MBONs in turn activates aversively reinforcing PPL1 DANs which feed back to encode a competing aversive odour memory. h, We propose that, after this extinction process, reduced CS+ drive to approach-directing MBONs (smaller blue circle) equals, and so neutralizes, the previously coded approach memory (smaller orange circle). i, Example of fly behaviour. Naive flies approach odours equally as a result of balanced drive to avoidance and approach MBON pathways. Reward-conditioned flies exhibit odour preference as a result of reduced CS+ drive to avoidance MBONs. Extinction restores the balance by reducing the CS+ drive to the approach MBONs. j, Sugar conditioning establishes enhanced CS− odour drive to γ2α′1 MBONs. k, During memory reactivation the CS− odour drives the γ2α′1 MBON which activates the MB-MV1 DAN that feeds back and releases dopamine within the same mushroom body compartment. This activity is required at the time of odour re-exposure to induce memory reconsolidation. l, CS− memory reactivation of the γ2α′1 MBON also activates rewarding DANs that innervate the tips of the horizontal lobes of the mushroom body, and that were earlier required for the formation of the original reward memory. The output of these rewarding DANs is required for a restricted period of time after odour exposure to reconsolidate memory.
Extended Data Figure 2 Extinction of reward memory requires negatively reinforcing dopaminergic neurons (related to Fig. 1).
a, 6 h reward memories for other odours (IAA and EB) can also be extinguished with two CS+ odour exposures 3 h after training (n ≥ 4). b, One or three odour exposures 3 h after training, varying the memory reactivation regimen of Fig. 1, abolish odour preference behaviour of trained flies measured 3 h later (n ≥ 6). c, Memory extinguished with two odour exposures at 3 h remains low 24 h after training (n = 8). Spontaneous recovery of the initial reward memory is not obvious with our current training and extinction protocols41. d, Two odour exposures, matching the memory reactivation regimen of Fig. 1, do not change the odour preference behaviour of naive flies measured 3 or 21 h later (n = 7). e, Blocking a reinforcement signal from rewarding R58E02-GAL4 DANs during retraining does not induce memory extinction (n ≥ 9). f, Permissive temperature control for Fig. 1c. No differences in CS+-directed extinction or approach behaviour following CS− exposure are apparent when the experiment in Fig. 1c is performed at permissive 23 °C throughout (n ≥ 7). g, Exposing flies to novel odours, IAA or EB, while MB504B-GAL4 PPL1 aversive DANs are blocked does not significantly impact 6 h memory performance (n ≥ 7). h, Blocking aversive PPL1 DANs during odour pre-exposure in naive flies does not attach a value to the pre-exposed odour (n ≥ 9).
Extended Data Figure 3 Reconsolidation of reward memory is triggered by CS− exposure and requires MB-MV1 dopaminergic neurons (related to Fig. 2).
a, Reward memories formed with other odours (IAA and EB) can also be rendered sensitive to cold-shock by reactivating them with CS− exposure 3 h after training (n = 9). b, Reward memories can also be made labile by reactivation 21 h after training (n = 10). c–e, Extinction of reward memory is insensitive to blocking small groups (<3 neurons per hemisphere) or individual classes of aversive PPL1 DANs during CS+-driven memory reactivation. Blocking MB-MP1 (n ≥ 10, c); MB-V1 (n ≥ 9, d) or PPL1-α3 and PPL1-α′3 (n ≥ 6, e) during CS− reactivation leaves 6 h memory performance unaltered. f, Manipulating the MB-MV1 DANs with the alternative driver R73F07-GAL4 during reactivation confirms a specific role in CS−-driven memory reconsolidation as seen with MB296B-GAL4 in Fig. 2c. Blocking R73F07-GAL4 neurons during CS+ reactivation does not affect reward memory extinction (n ≥ 14). g, Blocking MB-MV1 DANs (MB296B-GAL4) 90 min after CS− exposure does not impair reconsolidation (n ≥ 12). h, Permissive temperature control for f and Fig. 2c. CS− reactivation at permissive temperature does not change 6 h approach memory performance (n ≥ 8). i, MB-MV1 neurons are not required to form a 3 h sugar-rewarded memory (n ≥ 8).
Extended Data Figure 4 Reward memory extinction requires V2 cluster MBONs that drive negatively reinforcing dopaminergic neurons (related to Fig. 3).
a, Blocking the GABAergic MVP2 MBONs (MB112C-GAL4) during CS−- or CS+-triggered memory reactivation does not significantly impact 6 h conditioned approach behaviour or CS+-driven extinction (n ≥ 8). b, Permissive temperature control for Fig. 3a. Presenting the CS+ exposure at 23 °C does not change the extinction of reward memory in V2 MBON MB052B-GAL4;UAS-shits1 flies (n ≥ 8). c, d, Light-triggered activation (red bar) of R65B09-LexA V2 MBONs (c) or R24H08-LexA V2 MBONs (d) evokes calcium responses in PPL1 DANs. For c and d, asterisks denote significant differences (P < 0.05) between pre- and post-activation responses. e, Sugar-reward training does not alter CS+ or CS− odour-evoked calcium responses in V2 cluster MBONs MB052B-GAL4 (n ≥ 11). Responses to CS−, CS+ and novel odour were measured in a section through the α2 region of the vertical mushroom body lobe (example traces, lower left panel). Calcium transients during CS− and CS+ re-exposure were normalized to responses recorded in the same preparation to novel odour (IAA). f, Sugar-reward training does not alter CS+ or CS− odour-evoked calcium responses in MB-V1 or MB-MP1 MB052B-GAL4 DANs (n ≥ 7). Responses to CS−, CS+ and novel odour were measured in a section through the α2 or γ1 region of the mushroom body (example traces, lower left panel). Calcium transients during CS− and CS+ re-exposure were normalized to responses recorded to novel odour (IAA) in the same preparation. Note that the order of CS+ and CS− odour presentation is reversed for MB-V1 and MB-MP1 experiments.
Extended Data Figure 5 The γ2α′1 MBONs orchestrate CS−-triggered reconsolidation (related to Fig. 4).
a, Blocking the cholinergic MBON-γ2α′1 (MB077C-GAL4) after CS− exposure does not impair memory reconsolidation (n ≥ 10). b, Permissive temperature control for Fig. 4b. No defect in 3 h memory performance is apparent when the entire experiment is conducted at permissive 23 °C (n ≥ 11).
Extended Data Figure 6 The expression patterns of all GAL4 and LexA lines used in this study (related to Figs 1–4).
Panels a–k show GFP expression driven by the relevant GAL4 (green), LexA-driven RFP expression in mushroom body Kenyon cells (red) and general neuropil stained with an antibody to the Bruchpilot presynaptic marker (blue). a, R58E02-GAL4 broadly labels rewarding DANs in the PAM cluster including PAM-α1, PAM-β1 (MVP1), PAM-β1ped, PAM-β2, PAM-β′1ap, PAM-β′1m, PAM-β′2a PAM-β′2m, PAM-β′2p, PAM-γ3, PAM-γ4<γ1γ2, PAM-γ4 and PAM-γ5. b, TH-GAL4 broadly labels DANs throughout the brain including all six mushroom-body-innervating PPL1 DANs: PPL1-γ1pedc (MB-MP1), PPL1-γ1, PPL1-γ2α′1 (MB-MV1), PPL1-α′2α2 (MB-V1), PPL1-α3 and PPL1-α′3. c, MB504B-GAL4 labels PPL1-γ1pedc (MB-MP1), PPL1-γ2α′1 (MB-MV1), PPL1-α′2α2 (MB-V1) and PPL1-α3. d, e, MB296B-GAL4 (d) and R73F07-GAL4 (e) label PPL1-γ2α′1 (MB-MV1) neurons. f, c061-GAL4;MBGAL80 labels PPL1-γ1pedc (MB-MP1). g, MB058B-GAL4 labels PPL1-α′2α2 (MB-V1). h, MB308B-GAL4 labels PPL1-α′3 and displays weak expression in PPL1-α3. i, MB122C-GAL4 labels MBON-γ1pedc>α/β (MVP2). j, MB052B-GAL4 labels MBON-α′1, MBON-α2sc (V2α), MBON-α2p3p, MBON-α′3ap (V2α′) and MBON-α′3m (V2α′). k, MB077C-GAL4 labels MBON-γ2α′1. Panels l–p show GFP expression driven by the relevant LexA (green) and general neuropil stained with an antibody to the Bruchpilot presynaptic marker (blue). l, R65B09-LexA labels MBON-α′1, MBON-α2sc (V2α), MBON-α2p3p, MBON-α′2 (V4), MBON-α′3ap (V2α′) and MBON-α′3m (V2α′). m, R71D08-LexA labels MBON-α2sc (V2α), MBON-α′3ap (V2α′) and MBON-α′3m (V2α′). n, R24H08-LexA labels MBON-α′1, MBON-α′3ap (V2α′) and MBON-α′3m (V2α′). o, R58E02-LexA labels PAM-α1, PAM-β1 (MVP1), PAM-β1ped, PAM-β2, PAM-β′1ap, PAM-β′1m, PAM-β′2a, PAM-β′2m, PAM-β′2p, PAM-γ3, PAM-γ4<γ1γ2, PAM-γ4 and PAM-γ5. p, R25D01-LexA labels MBON-γ2α′1.
Supplementary information
Supplementary Table 1
This file contains details of sample numbers and statistics for Figures 1-4. (XLSX 30 kb)
Supplementary Table 2
This file contains details of sample numbers and statistics for Extended Data Figures 2-5. (XLSX 31 kb)
Rights and permissions
About this article
Cite this article
Felsenberg, J., Barnstedt, O., Cognigni, P. et al. Re-evaluation of learned information in Drosophila. Nature 544, 240–244 (2017). https://doi.org/10.1038/nature21716
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21716
This article is cited by
-
A hippocampus-accumbens code guides goal-directed appetitive behavior
Nature Communications (2024)
-
Dopaminergic systems create reward seeking despite adverse consequences
Nature (2023)
-
How flies remember a rich experience from its individual components
Nature (2023)
-
Forgetting as a form of adaptive engram cell plasticity
Nature Reviews Neuroscience (2022)
-
Acetylcholine deficit causes dysfunctional inhibitory control in an aging-dependent manner
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.