Abstract
Adiponectin receptors (ADIPORs) are integral membrane proteins that control glucose and lipid metabolism by mediating, at least in part, a cellular ceramidase activity1 that catalyses the hydrolysis of ceramide to produce sphingosine and a free fatty acid (FFA). The crystal structures of the two receptor subtypes, ADIPOR1 and ADIPOR2, show a similar overall seven-transmembrane-domain architecture with large unoccupied cavities and a zinc binding site within the seven transmembrane domain2. However, the molecular mechanisms by which ADIPORs function are not known. Here we describe the crystal structure of ADIPOR2 bound to a FFA molecule and show that ADIPOR2 possesses intrinsic basal ceramidase activity that is enhanced by adiponectin. We also identify a ceramide binding pose and propose a possible mechanism for the hydrolytic activity of ADIPOR2 using computational approaches. In molecular dynamics simulations, the side chains of residues coordinating the zinc rearrange quickly to promote the nucleophilic attack of a zinc-bound hydroxide ion onto the ceramide amide carbonyl. Furthermore, we present a revised ADIPOR1 crystal structure exhibiting a seven-transmembrane-domain architecture that is clearly distinct from that of ADIPOR2. In this structure, no FFA is observed and the ceramide binding pocket and putative zinc catalytic site are exposed to the inner membrane leaflet. ADIPOR1 also possesses intrinsic ceramidase activity, so we suspect that the two distinct structures may represent key steps in the enzymatic activity of ADIPORs. The ceramidase activity is low, however, and further studies will be required to characterize fully the enzymatic parameters and substrate specificity of ADIPORs. These insights into ADIPOR function will enable the structure-based design of potent modulators of these clinically relevant enzymes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Holland, W. L. et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat. Med. 17, 55–63 (2011)
Tanabe, H. et al. Crystal structures of the human adiponectin receptors. Nature 520, 312–316 (2015)
Scherer, P. E., Williams, S., Fogliano, M., Baldini, G. & Lodish, H. F. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 270, 26746–26749 (1995)
Berg, A. H., Combs, T. P., Du, X., Brownlee, M. & Scherer, P. E. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 7, 947–953 (2001)
Fruebis, J . et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc. Natl Acad. Sci. USA 98, 2005–2010 (2001)
Arita, Y. et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 257, 79–83 (1999)
Hotta, K. et al. Circulating concentrations of the adipocyte protein adiponectin are decreased in parallel with reduced insulin sensitivity during the progression to type 2 diabetes in rhesus monkeys. Diabetes 50, 1126–1133 (2001)
Yamauchi, T. et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 7, 941–946 (2001)
Yamauchi, T. et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423, 762–769 (2003)
Tang, Y. T. et al. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J. Mol. Evol. 61, 372–380 (2005)
Marheineke, K., Grünewald, S., Christie, W. & Reiländer, H. Lipid composition of Spodoptera frugiperda (Sf9) and Trichoplusia ni (Tn) insect cells used for baculovirus infection. FEBS Lett. 441, 49–52 (1998)
Airola, M. V. et al. Structural basis for ceramide recognition and hydrolysis by human neutral ceramidase. Structure 23, 1482–1491 (2015)
Villa, N. Y. et al. Sphingolipids function as downstream effectors of a fungal PAQR. Mol. Pharmacol. 75, 866–875 (2009)
Langosch, D., Scharnagl, C., Steiner, H. & Lemberg, M. K. Understanding intramembrane proteolysis: from protein dynamics to reaction kinetics. Trends Biochem. Sci. 40, 318–327 (2015)
Kamp, F. et al. Intramembrane proteolysis of β-amyloid precursor protein by γ-secretase is an unusually slow process. Biophys. J. 108, 1229–1237 (2015)
Pei, J., Millay, D. P., Olson, E. N. & Grishin, N. V. CREST—a large and diverse superfamily of putative transmembrane hydrolases. Biol. Direct 6, 37 (2011)
Holland, W. L., Xia, J. Y., Johnson, J. A. & Scherer, P. E. Inducible overexpression of adiponectin receptors highlight the roles of adiponectin-induced ceramidase signaling in lipid and glucose homeostasis. Mol. Metab. 6, 267–275 (2017)
Hernick, M. & Fierke, C. A. Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 433, 71–84 (2005)
Gilmartin, A. A. et al. High-level secretion of recombinant monomeric murine and human single-chain Fv antibodies from Drosophila S2 cells. Protein Eng. Des. Sel. 25, 59–66 (2012)
Johansson, D. X., Krey, T. & Andersson, O. Production of recombinant antibodies in Drosophila melanogaster S2 cells. Methods Mol. Biol. 907, 359–370 (2012)
Krey, T. et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6, e1000762 (2010)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protocols 4, 706–731 (2009)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Winter, G., Lobley, C. M. & Prince, S. M. Decision making in xia2. Acta Crystallogr. D 69, 1260–1273 (2013)
Foadi, J. et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr. D 69, 1617–1632 (2013)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Bricogne G . et al. BUSTER Version X.Y.Z. (Global Phasing Ltd., Cambridge, UK, 2016)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013)
Saied, E. M., Banhart, S., Bürkle, S. E., Heuer, D. & Arenz, C. A series of ceramide analogs modified at the 1-position with potent activity against the intracellular growth of Chlamydia trachomatis. Future Med. Chem. 7, 1971–1980 (2015)
Korb, O., Stützle, T. & Exner, T. E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 49, 84–96 (2009)
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H. J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–367 (2005)
Mashiach, E., Schneidman-Duhovny, D., Andrusier, N., Nussinov, R. & Wolfson, H. J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 36, W229–232 (2008)
Grosdidier, A., Zoete, V. & Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–277 (2011)
Banks, J. L. et al. Integrated modeling program, applied chemical theory (IMPACT). J. Comput. Chem. 26, 1752–1780 (2005)
Friesner, R. A . et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004)
Halgren, T. A . et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759 (2004)
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006)
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014)
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
Venable, R. M. et al. CHARMM all-atom additive force field for sphingomyelin: elucidation of hydrogen bonding and of positive curvature. Biophys. J. 107, 134–145 (2014)
Vanommeslaeghe, K. et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010)
Steinbach, P. J. & Brooks, B. R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 15, 667–683 (1994)
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995)
Parrinello, M. & Rahman, A. Polymorphic transitions in single-crystals — a new molecular-dynamics method. J. Appl. Phys. 52, 7182–7190 (1981)
Hoover, W. G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31, 1695–1697 (1985)
Nose, S. A molecular-dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984)
Acknowledgements
We thank C. Mueller-Dieckmann and U. Zander at the European Synchrotron Radiation Facility (ESRF) for assistance in using beamline ID30B. We acknowledge the ESRF for provision of synchrotron radiation facilities via SSX Block Allocation Group beamtime. We thank R. Joosten and A. Perrakis from the PDB REDO server for help with ADIPOR1 data re-analysis and F. Rey from the Structural Virology Unit, Institut Pasteur for S2 cells and an expression vector for scFv. This project received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement 647687).
Author information
Authors and Affiliations
Contributions
I.V.-B. and R.S. expressed, purified, characterized and crystallized receptor and scFv preparations with the help of P.R., G.B., M.F. and C.B. I.V.-B. and C.L. collected data with the help of F.H. C.L. performed the computational studies with help from L.D.C. I.V.-B. and C.L. solved and refined the structures. R.S. prepared the figures with the help of I.V.-B. and C.L. E.M.S. synthesized ceramides of different chain lengths and performed UPLC–MS analysis of the ceramide cleavage reactions. C.A. supervised E.M.S. All authors contributed to the manuscript preparation. S.G. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Veprintsev and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
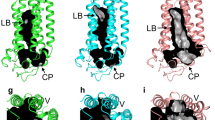
Extended Data Figure 1 Comparison of the three ADIPOR2 structures.
a, Comparison of the original (top) and revised (bottom) ADIPOR2 crystal structures. The modified sections and the additional molecules modelled in the revised structures are highlighted in red. b, Overall view of ADIPOR2–scFv crystal structures from within the membrane plane. The heavy and light chain variable regions (VH and VL) are coloured dark and light grey, respectively. Oleic acid (FA C18:1) is shown as sticks with C atoms displayed as spheres and coloured according to element: carbon, blue; oxygen, red. c, 2Fo−Fc (dark grey) and Fo−Fc (light grey) density maps used to position oleic acid contoured at 1σ and 2.5σ, respectively. The density provided sufficient features to reliably position a free fatty acid in all structures. However, we could not make a clear distinction between an oleate (C18:1) and a stearate (C18:0) but decided to model an oleate because it is present in greater amounts than stearate in insect cells and the statistics were marginally better. d, Hydrophobic binding pocket of the oleic acid within ADIPOR2 displayed as transparent blue surface. Residues forming the pocket are shown as sticks. e, 2Fo−Fc electron density around the zinc binding site contoured at 1σ in S1, S2 and S3 crystal structures viewed from the intracellular side. The electron density reveals distinct positions of the carboxylic acid moiety and of the tentatively assigned water molecules resulting in the different apparent coordination geometries of the zinc ion. S1, S2 and S3 crystal structures are shown as cartoons and coloured dark yellow, light blue and pink, respectively. The zinc ion is represented as an orange sphere. Residues participating in zinc coordination and carboxylic acid moiety are shown as sticks with oxygen and nitrogen atoms coloured red and dark blue, respectively. Oleic acid is shown as sticks with C atoms displayed as spheres and coloured according to element: carbon, blue; oxygen, red. Water molecules are shown as red spheres.
Extended Data Figure 2 Features of the ADIPOR2 continuous cavity.
a, Extra electron density (2Fo−Fc at 1σ) in the tunnel between TM5 and TM6 assigned to monoolein (rac-glycerol 1-monooleate) as it is the most concentrated component in the crystallization sample and probably binds this region with its oleate C18:1 moiety. We cannot rule out the possibility that the density originates from another molecule containing a long aliphatic chain. In both cases, the extra density suggests that this opening may play a role in ADIPOR2 function. The occupancy of the glycerol moiety and of the first four carbons from the ester group were, however, set to 0 during further refinement in the absence of a significant electron density as indicated by the 2Fo−Fc map contoured at 1σ. Monoolein and oleic acid are shown as sticks with C atoms displayed as spheres and coloured according to element: carbon, yellow and blue, respectively for monoolein and oleic acid; oxygen, red. Tentatively assigned water molecules (red spheres) in an extra pocket close to the zinc site (b), and in the intracellular cavity and the intracellular opening (c). The extra pocket close to the zinc site is separated from the FFA molecule by F351TM7 and I223TM3 and is filled with water molecules. This cavity might be a reservoir of water molecules for the hydrolytic activity but this hypothesis remains to be proven. The intracellular pocket is split into the intracellular cavity (blue broken line) and the intracellular opening (yellow broken line) by residues D117Nter and Y205ICL1. Computational studies suggest that the sphingosine diffuses out from the receptor through the intracellular opening. The role of the intracellular cavity is hard to predict at this time. d, Residues forming the intracellular cavity (blue) and opening (yellow).
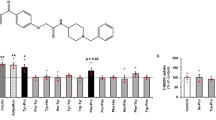
Extended Data Figure 3 Biochemical analyses of ceramide binding to ADIPOR1 and ADIPOR2 and sphingosine formation.
a, b, A specific fluorescent signal for ADIPOR2 (a) or ADIPOR1 (b) incubated with NBD–ceramide was observed by FSEC at the elution volume of the receptor (blue broken line). In both cases, the two main peaks in the SEC absorbance traces could correspond to at least two receptor populations but only the one corresponding to the smaller peak on the right can bind to NBD–ceramide, suggesting that part of the protein is either not functional or in a conformation that cannot accommodate the substrate. The presented figure is representative of two experiments performed with two independent receptor preparations. c, d, Sphingosine detected by LC–MS analysis revealed ADIPOR1 ceramidase activity and adiponectin stimulation (around 25-fold increase over basal). Relative sphingosine (c) and detected sphingosine (d) values are represented as the mean ± s.d. of three independent measurements. e, Mass spectrum for the extracted ion peak (retention time of 2.27 min) of ADIPOR2 with ceramide–C24 sample, and the d-erythro-sphingosine (d18:1) standard sample. f, Representative LC–MS analysis with an extracted ion chromatogram (m/z from 299.7 to 300.7) of the d-erythro-sphingosine (d18:1) standard sample (left) and of ceramide C24 with n-dodecyl-β-d-maltopyranoside (DDM) and cholesteryl hemisuccinate (CHS) sample (right) in which no signals for sphingosine were detected. g, Representative LC–MS analysis with an extracted ion chromatogram (m/z = 299.7–300.7) revealing the formation of the sphingoid base sphingosine (m/z = 300.3, retention time 2.26 min) from the enzymatic reaction between ADIPOR2 and ceramides of different chain lengths: ceramide C6, ceramide C18 and ceramide C24. The bottom panel represents untreated samples of ADIPOR2. h, Representative LC–MS analysis with an extracted ion chromatogram (m/z = 299.7–300.7) revealing the formation of the sphingoid base sphingosine (m/z = 300.3, retention time 2.27 min) from the enzymatic reaction between ADIPOR1 and ceramides of different chain lengths: ceramide C6, ceramide C18 and ceramide C24. The bottom panel represents the untreated samples of ADIPOR1.
Extended Data Figure 4 MDS of ADIPOR2 substrate and product complexes.
a, Comparison of the S1 crystal structure (PDB 5LX9) with the starting model used for MDS of the fatty acid and sphingosine system. A close-up of the active site is shown in the inset. b, d, Calculated r.m.s.d. and distances between indicated residues and zinc during MDS performed with sphingosine and FA (C16:0) (b) or with sphingosine and FA (C18:1) (d). In both cases, the sphingosine leaves the active site within the time scale of the MDS and moves towards the cytoplasm. The zinc coordination sphere remains as observed in the crystal structures during the C16:0 MDS (that is, H202, H348, and H352 interact with the zinc ion), whereas in the C18:1 MDS some differences are observed. In the first 150 ns, S198 interacts with the zinc along with H202, H348, and H352, while D219 is involved in a salt bridge with the sphingosine amine. After 150 ns, the D219–sphingosine interaction is broken and D219 replaces S198 in the zinc coordination sphere. These differences are likely to arise from the destabilization of the active site by the FA and sphingosine, which probably requires longer time scales to return to equilibrium, as well as the strong tendency of MDS to remain stuck in local energy minima. c, Snapshots of the active site extracted from MDS in the presence of the sphingosine and the FA (C16:0) at different times showing that the zinc binding site remained in the configuration observed in the crystal structures (except that the zinc adopted an octahedral geometry by interacting with three water molecules). e, Snapshots of the active site extracted from MDS in the presence of the sphingosine and FA (C18:1) at different times. In both C16:0 and C18:1 trajectories, the FA carboxylate forms a salt bridge with R278 side chain, which is also observed in the S2 structure. f, Snapshots of the fatty acid and sphingosine taken every 10 ns along the 460-ns MDS trajectory, highlighting the movements of the sphingosine inside the receptor. The fatty acid and sphingosine are represented as blue and green lines, respectively. The zinc atom is shown as an orange sphere and the receptor is shown in cartoon representation. The carboxylic acid carbon (C1) of the fatty acid and the nitrogen atom of the sphingosine are shown in spheres and coloured using blue-to-orange and green-to-red gradients to help visualize their motion over simulation time. g, C16:0 ceramide top-scoring docking pose, reminiscent of the C18:1 pose. h, Calculated r.m.s.d. and distances between indicated residues and zinc (h) during MDS performed with the C16:0 ceramide revealing a behaviour similar to C18:1 ceramide. Inset in h highlights changes happening at the very beginning of the MDS. Because only the ligand is flexible during docking, and ADIPOR2 receptor was crystallized in a state that corresponds to the product state of the reaction (step four in the proposed mechanism), our interpretation is that the initial relaxation of the system represents the structural adaptation of the receptor to the presence of the substrate (induced fit back to step 1). We suspect that the movements of the receptor are fast because of the presence of the substrate in the binding pocket which is in the product state conformation thus constituting a perturbation of the system. i, Snapshots of the active site extracted from MDS in the presence of C16:0 ceramide at different simulation times. The C16:0 ceramide and zinc-coordinating residues sampled conformations similar to those observed for C18:1 ceramide. At late time points in the simulation, the ceramide moved slightly away from the zinc binding site, probably as a consequence of the inability of MDS to simulate the destruction and creation of covalent bonds.
Extended Data Figure 5 Proposed catalytic mechanism of ADIPOR2 ceramidase activity and docking calculations.
a, On the basis of the interactions with ceramide and conformational changes of the zinc active site observed in MDS, as well as the zinc-coordinated FFA carboxyl oxygen seen in the crystal structures, we propose a general acid-base catalysis mechanism for the hydrolysis of the amide bond by ADIPOR2. In this mechanism, which is similar to that proposed for neutral ceramidase (ref. 12 in the main text), the zinc ion activates a water molecule for nucleophilic attack of the amide carbon (1). Y220TM3, R278TM5 and Y328TM6 side chains polarize the amide carbonyl and stabilize the oxyanion formed in the tetrahedral transition state (2). H348TM7 serves as a general base for proton extraction from water (1) and subsequently acts as a general acid to transfer this proton to the nitrogen of ceramide during or immediately after amide bond cleavage (3). The active site rearranges following the hydrolysis reaction to yield the product state-associated zinc coordination sphere observed in the crystal structures and MDS (4). In this study, we decided to perform docking and simulations with ceramides and FFA presenting two different acyl chain lengths (C16:0, C18:1) as we anticipated that chain length may not have a major impact on the observed mechanism and to compare our results with the study of ceramide binding to neutral ceramidase12, in which docking calculations were performed with C16:0. b, The top scoring C18:1 FA docking pose obtained using PLANTS (shown as green sticks) is superimposed on the crystallographically observed fatty acid taken from the revised ADIPOR2 structure (5LWY) (blue balls and sticks representation). c, Comparison of the top-scoring C18:1 ceramide docking poses obtained using three different docking programs (PLANTS, Patchdock/Firedock and Glide). The ligands are shown as sticks with hydrogens omitted for clarity. The top-scoring pose from PLANTS and Patchdock/Firedock are very similar, while the glide pose is slightly shifted towards the cytoplasm and a significant portion of the sphingosine moiety is exposed to the cytoplasm. In all three cases, the ceramide carbonyl contacts the Y328 side chain. d, Comparison of the C18:1 and C16:0 ceramide top-scoring docking poses obtained using PLANTS. The ADIPOR2 receptor is shown as semi-transparent cartoon and surface, and the insets highlight the position of the sphingosine moiety relative to the intracellular surface of the receptor.
Extended Data Figure 6 The putative catalytic residues and the substrate binding pocket are highly conserved in the PAQR family.
a, View of conserved residues around the zinc ion (orange sphere) from the intracellular side. The evolutionary analysis performed by Consurf server50 reveals that residues H202, H348, H352, and D219 (shown as sticks) coordinating the zinc ion in ADIPOR2 are strictly conserved in the entire PAQR family. S198, which is potentially involved in ceramide hydrolysis, is also strictly conserved within members of the human PAQR family. The receptor is shown as cartoon and coloured using the Consurf colour scale. Oxygen and nitrogen atoms are coloured red and dark blue, respectively. b, The conservation of the internal cavity within PAQR family viewed from within the membrane in two orientations obtained by a 180° rotation. The cavity is represented in surface (cavity mode 1) and coloured using the Consurf colour scale. These data strongly suggest that all members of the PAQR family may have ceramidase activity. c, Sequence alignment of the 11 members of the PAQR family coloured using the Consurf colour scale. It is important to note that ADIPORs also share some homology with alkaline ceramidases, further reinforcing the experimental evidence found in this study.
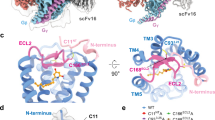
Extended Data Figure 7 Corrected electron density and ADIPOR1 TM5 positions and crystal lattice packing of ADIPOR1–scFv and ADIPOR2–scFv.
a–c, 2Fo−Fc (blue mesh) and Fo−Fc (green mesh) electron density maps around TM5 are contoured at 1σ and 2.5σ, respectively, in the initial ADIPOR1 structure (PBD 3WXV) fetched from the Electron Density Server (a), the ADIPOR1 structure after modelling in strong positive difference map peaks at ~13σ and ~6σ with two sulfate ions, respectively (b) and the final revised ADIPOR1 structure (c). ADIPOR1 is shown as cartoon and coloured in light grey with TM5 highlighted in black. The zinc ion is shown as an orange sphere. d–f, Lattice packing of ADIPOR2–scFv crystals viewed within the membrane plane (d, e) and from the extracellular side (f). ADIPOR2 TM5 (red) does not make any crystal contacts with the bound scFv or the symmetry related molecules. g–i, Lattice packing of ADIPOR1–scFv crystals viewed within the membrane plane (g, h) and from the extracellular side (i). TM5 (red) of ADIPOR1 (R1-a) makes contact with TM1 and the N-terminal short helix (helix 0) (both in blue) from another symmetry-related ADIPOR1 molecule (R1-b). j, k, Closer view of the interaction of ADIPOR1 TM5 with TM1 and helix 0 from the symmetry-related ADIPOR1 molecule. At the top, TM5 is stabilized by hydrophobic contact between I287 and F125 of the symmetry-related helix 0 as well as hydrogen bonds between the main chain carbonyl of A288 in TM5 and R122 in helix 0 as indicated by the black dashed line (j). At the bottom of TM5, Q265 interacts with R158 of the symmetry-related receptor molecule ICL1 as indicated by the black dashed line (k). In addition, F271, L272 and L276 make hydrophobic contacts with symmetry-related TM1 residues L157, I153, F150 and L149. The interacting residues are displayed as sticks and coloured green for TM5 and yellow for TM1. ADIPOR1, ADIPOR2, VH and VL are coloured pale green, wheat, dark grey and light grey, respectively. In both ADIPOR1–scFv and ADIPOR2–scFv, scFv molecules are contributing the most to the crystal lattice formation. Regarding the contacts between just the receptor molecules, in ADIPOR2 the packing is also mediated by TM4 of two symmetry-related molecules.
Extended Data Figure 8 A broken N terminus–TM5 interaction between the closed (ADIPOR2) and open (ADIPOR1) structures.
a, b, The polar interaction between R275TM5 and D117N-term of ADIPOR2 (a) is broken in the open ADIPOR1 structure with the corresponding R264TM5 shifted away and the D106N-term side chain repositioned to interact with Y194 (b). ADIPOR1 and ADIPOR2 are shown as cartoons and coloured light green and dark yellow, respectively. R275, D117, R264, D106 and Y194 are shown as sticks. The zinc ion is represented as an orange sphere. Residues coordinating the zinc ion are shown as lines. Oxygen and nitrogen atoms are coloured red and dark blue, respectively.
Supplementary information
Supplementary Information
This file contains Supplementary Text, additional references and Supplementary Tables 1-3. (PDF 195 kb)
Rights and permissions
About this article
Cite this article
Vasiliauskaité-Brooks, I., Sounier, R., Rochaix, P. et al. Structural insights into adiponectin receptors suggest ceramidase activity. Nature 544, 120–123 (2017). https://doi.org/10.1038/nature21714
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21714
This article is cited by
-
Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1
Cell Discovery (2024)
-
Sphingolipids: drivers of cardiac fibrosis and atrial fibrillation
Journal of Molecular Medicine (2024)
-
PAQR8 promotes breast cancer recurrence and confers resistance to multiple therapies
Breast Cancer Research (2023)
-
Expression of Myomaker and Myomerger in myofibers causes muscle pathology
Skeletal Muscle (2023)
-
Structure, dynamics and transferability of the metal-dependent polyhistidine tetramerization motif TetrHis for single-chain Fv antibodies
Communications Chemistry (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.