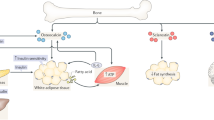
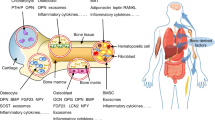
Abstract
Bone has recently emerged as a pleiotropic endocrine organ that secretes at least two hormones, FGF23 and osteocalcin, which regulate kidney function and glucose homeostasis, respectively. These findings have raised the question of whether other bone-derived hormones exist and what their potential functions are. Here we identify, through molecular and genetic analyses in mice, lipocalin 2 (LCN2) as an osteoblast-enriched, secreted protein. Loss- and gain-of-function experiments in mice demonstrate that osteoblast-derived LCN2 maintains glucose homeostasis by inducing insulin secretion and improves glucose tolerance and insulin sensitivity. In addition, osteoblast-derived LCN2 inhibits food intake. LCN2 crosses the blood–brain barrier, binds to the melanocortin 4 receptor (MC4R) in the paraventricular and ventromedial neurons of the hypothalamus and activates an MC4R-dependent anorexigenic (appetite-suppressing) pathway. These results identify LCN2 as a bone-derived hormone with metabolic regulatory effects, which suppresses appetite in a MC4R-dependent manner, and show that the control of appetite is an endocrine function of bone.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Friedman, J. M. & Halaas, J. L. Leptin and the regulation of body weight in mammals. Nature 395, 763–770 (1998)
Brüning, J. C. et al. Role of brain insulin receptor in control of body weight and reproduction. Science 289, 2122–2125 (2000)
Drucker, D. J. The biology of incretin hormones. Cell Metab. 3, 153–165 (2006)
Choudhury, A. I. et al. The role of insulin receptor substrate 2 in hypothalamic and β cell function. J. Clin. Invest. 115, 940–950 (2005)
Batterham, R. L. et al. Critical role for peptide YY in protein-mediated satiation and body-weight regulation. Cell Metab. 4, 223–233 (2006)
Coll, A. P., Farooqi, I. S. & O’Rahilly, S. The hormonal control of food intake. Cell 129, 251–262 (2007)
Lee, N. K. et al. Endocrine regulation of energy metabolism by the skeleton. Cell 130, 456–469 (2007)
Ferron, M. et al. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell 142, 296–308 (2010)
Fulzele, K. et al. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell 142, 309–319 (2010)
Rached, M. T. et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J. Clin. Invest. 120, 357–368 (2010)
Bhattacharyya, N., Chong, W. H., Gafni, R. I. & Collins, M. T. Fibroblast growth factor 23: state of the field and future directions. Trends Endocrinol. Metab. 23, 610–618 (2012)
Yoshikawa, Y. et al. Genetic evidence points to an osteocalcin-independent influence of osteoblasts on energy metabolism. J. Bone Miner. Res. 26, 2012–2025 (2011)
Huszar, D. et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88, 131–141 (1997)
Butler, A. A. & Cone, R. D. The melanocortin receptors: lessons from knockout models. Neuropeptides 36, 77–84 (2002)
Balthasar, N. et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell 123, 493–505 (2005)
Liu, H. et al. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J. Neurosci. 23, 7143–7154 (2003)
Ste Marie, L., Miura, G. I., Marsh, D. J., Yagaloff, K. & Palmiter, R. D. A metabolic defect promotes obesity in mice lacking melanocortin-4 receptors. Proc. Natl Acad. Sci. USA 97, 12339–12344 (2000)
Coll, A. P., Farooqi, I. S., Challis, B. G., Yeo, G. S. & O’Rahilly, S. Proopiomelanocortin and energy balance: insights from human and murine genetics. J. Clin. Endocrinol. Metab. 89, 2557–2562 (2004)
Hinney, A. et al. Prevalence, spectrum, and functional characterization of melanocortin-4 receptor gene mutations in a representative population-based sample and obese adults from Germany. J. Clin. Endocrinol. Metab. 91, 1761–1769 (2006)
Larsen, L. H. et al. Prevalence of mutations and functional analyses of melanocortin 4 receptor variants identified among 750 men with juvenile-onset obesity. J. Clin. Endocrinol. Metab. 90, 219–224 (2005)
Nix, M. A. et al. Molecular and functional analysis of human β-defensin 3 action at melanocortin receptors. Chem. Biol. 20, 784–795 (2013)
Yan, Q. W. et al. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes 56, 2533–2540 (2007)
Lin, Y. et al. Hyperglycemia-induced production of acute phase reactants in adipose tissue. J. Biol. Chem. 276, 42077–42083 (2001)
Soukas, A., Cohen, P., Socci, N. D. & Friedman, J. M. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 14, 963–980 (2000)
Dacquin, R., Starbuck, M., Schinke, T. & Karsenty, G. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dyn. 224, 245–251 (2002)
Kalajzic, Z. et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone 31, 654–660 (2002)
Qiang, G. et al. Lipodystrophy and severe metabolic dysfunction in mice with adipose tissue-specific insulin receptor ablation. Mol. Metab. 5, 480–490 (2016)
Scheller, E. L., Cawthorn, W. P., Burr, A. A., Horowitz, M. C. & MacDougald, O. A. Marrow adipose tissue: trimming the fat. Trends Endocrinol. Metab. 27, 392–403 (2016)
Zhang, M. et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 277, 44005–44012 (2002)
Coleman, D. L. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia 14, 141–148 (1978)
Kelly, E., Bailey, C. P. & Henderson, G. Agonist-selective mechanisms of GPCR desensitization. Br. J. Pharmacol. 153 (Suppl 1), S379–S388 (2008)
Gainetdinov, R. R., Premont, R. T., Bohn, L. M., Lefkowitz, R. J. & Caron, M. G. Desensitization of G protein-coupled receptors and neuronal functions. Annu. Rev. Neurosci. 27, 107–144 (2004)
Garfield, A. S. et al. A neural basis for melanocortin-4 receptor-regulated appetite. Nat. Neurosci. 18, 863–871 (2015)
Myers, M. G., Jr & Olson, D. P. Central nervous system control of metabolism. Nature 491, 357–363 (2012)
Cone, R. D. Anatomy and regulation of the central melanocortin system. Nat. Neurosci. 8, 571–578 (2005)
Berglund, E. D. et al. Melanocortin 4 receptors in autonomic neurons regulate thermogenesis and glycemia. Nat. Neurosci. 17, 911–913 (2014)
Kublaoui, B. M., Holder, J. L., Jr, Gemelli, T. & Zinn, A. R. Sim1 haploinsufficiency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons. Mol. Endocrinol. 20, 2483–2492 (2006)
Law, I. K . et al. Lipocalin-2 deficiency attenuates insulin resistance associated with aging and obesity. Diabetes 59, 872–882 (2010)
Guo, H.,et al. Lipocalin-2 deficiency impairs thermogenesis and potentiates diet-induced insulin resistance in mice. Diabetes 59, 1376–1385 (2010)
Eguchi, J. et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13, 249–259 (2011)
Paragas, N. et al. The Ngal reporter mouse detects the response of the kidney to injury in real time. Nat. Med. 17, 216–222 (2011)
Parton, L. E. et al. Glucose sensing by POMC neurons regulates glucose homeostasis and is impaired in obesity. Nature 449, 228–232 (2007)
van den Pol, A. N. et al. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J. Neurosci. 29, 4622–4639 (2009)
DeFronzo, R. A., Tobin, J. D. & Andres, R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am. J. Physiol. 237, E214–E223 (1979)
Kode, A. et al. FOXO1 orchestrates the bone-suppressing function of gut-derived serotonin. J. Clin. Invest. 122, 3490–3503 (2012)
Sohn, J. W. et al. Serotonin 2C receptor activates a distinct population of arcuate pro-opiomelanocortin neurons via TRPC channels. Neuron 71, 488–497 (2011)
Williams, K. W. et al. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 20, 471–482 (2014)
Mellon, P. L. et al. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron 5, 1–10 (1990)
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995)
Cohen J. in Statistical Power Analysis for the Behavioral Sciences 24–27 (Lawrence Erlbaum Associates, 1988)
Acknowledgements
The authors are grateful to A. Tarasenco for technical assistance, the Histology and Metabolic Unit facility of the Diabetes and Endocrinology Research Center (DERC, NIDDK DK063608-07), R. Tomaino of the Taplin Mass Spectrometry Facility, Harvard Medical School and the Albert Einstein DRTC Animal Physiology Core (NIDDK DK20541). This work was supported by the National Institutes of Health R01AR054447, P01AG032959 and R01AR055931 to S.K., R01DK100699 to K.W.W., R01DK52431 to R.L.L. and W.C., P30DK26687 to R.L.L. and W.C., P30DK063608 to R.L.L. and W.C., NOVO Nordisk to S.K and by the T32 Training Grant DK07328 to S.S.
Author information
Authors and Affiliations
Contributions
I.M. and S.K. initiated the study, designed experiments and analysed data. I.M. and S.S. performed experiments and analysed data. N.L., J.-M.L. and A.M. helped with ICV infusions and bone phenotyping. H.Z. performed hyperglycemic clamps. R.A.F. analysed microarray data. J.B. provided Lcn2-reporter mice. P.L., L.D., R.L.L. and W.C. provided plasma samples of patients with MC4R mutations. M.R. and T.N. provided serum samples from men with type 2 diabetes. Z.H., Y.H. and K.W.W. performed the electrophysiology experiments. L.M.Z. and J.E.P. discussed data and manuscript. I.M. and S.K. wrote the manuscript. S.K. directed the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks M. Myers, R. Palmiter, M. Zaidi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Identification of Lcn2 as an osteoblast-enriched gene and generation of mice lacking Lcn2 in osteoblasts (Lcn2osb−/−) and adipocytes (Lcn2fat−/−).
a, Real-time PCR analysis of Lcn2 expression levels in the indicated tissues from wild-type mice. b, Lcn2, Bglap and Alp expression levels in differentiating osteoblasts (day 0–day 15). c, Lack of Lcn2 expression in indicated tissues of the Lcn2–mCherry-reporter mouse. Scale bars, 40 μm. d, Targeting strategy used to generate a floxed allele of Lcn2. The targeting vector, which contains loxP sites within introns 2 and 6, is designed to delete a 1.9 kb genomic fragment containing Lcn2 exons 3−6. Location of probes used for Southern blotting (5′ and 3′) and primers used for PCR to detect the floxed (a and b) and mutant allele (c and d) are indicated. e, Southern-blot analysis on DNA from targeted embryonic stem (ES) cells and mice from F1 and F2 generation showing germline transmission of the mutated allele. f, i, Detection of Lcn2 floxed (primers a–b) and mutant allele, Δflox, (primers c–d) in genomic DNA isolated from tissues of Lcn2osb−/− (f) and Lcn2fat−/− (i) mice. Lcn2osb−/− mice showed 60% recombination efficiency and 55% decrease in Lcn2 expression specifically in bone, but not in other tissues. Lcn2fat−/− mice showed 76% recombination efficiency in fat, which led to 65% and 50% decrease in Lcn2 expression in white and brown fat, respectively. g, j, Serum levels of LCN2 are decreased by 67% in Lcn2osb−/− mice compared to their Lcn2fl/fl littermates (g), whereas levels are decreased by 27% in Lcn2fat−/− mice compared to their Lcn2fl/fl littermates (j). h, k, Tissue expression of Lcn2 in Lcn2osb−/− and their Lcn2fl/fl littermates (h) and Lcn2fat−/− and their Lcn2fl/fl littermates (k). Data are mean ± s.e.m.; n = 12 (a) and n = 9 (g, h, j, k) mice per group. Results are representative of three independent experiments. *P < 0.05 (Student’s t-test).
Extended Data Figure 2 Osteoblast-specific activation of Cre in Col1a1-Cre and Bglap-Cre mice.
a, b, Tandem mass spectrum of an eluted peptide fragment of bone-derived (a) and fat-derived (b) LCN2. c, Specific expression of Cre in osteoblasts, but not any other tissues in mT/mG::Col1a1-Cre mice and in mT/mG::Bglap-Cre mice. Scale bars, 100 μm. Results are representative of three independent experiments.
Extended Data Figure 3 LCN2 regulates glucose homeostasis through its expression in osteoblasts.
a, b, Glucose infusion rate (a) and serum insulin levels (b) during hyperglycaemic clamp. c, Insulin staining and islet histomorphometry of pancreatic sections. d, Ki67 immunostaining and histomorphometric analysis of Ki67-immunoreactive cells in pancreatic islets (arrows indicate Ki67-positive cells). e, Histomorphometric analysis of TUNEL-positive β-cells in the pancreas. f, Expression of insulin genes, cell cycle genes and β-cell dedifferentiation markers in pancreatic islets. g, Insulin and glucagon immunostaining and histomorphometric analysis of insulin- and glucagon-immunoreactive cells in pancreatic islets of Lcn2osb−/− mice and their Lcn2fl/fl littermates. h, Insulin secretion in pancreatic islets treated with increasing doses of LCN2. i–p, Glucose tolerance (i), insulin tolerance (j) and glucose-stimulated insulin secretion (k), random-fed serum-insulin levels (l), fat pad mass (m), fat mass (n), food intake (o) and body weight (p) are not altered in Col1a1-Cre mice compared to their wild-type littermates. q, Expression levels of Cebpa, Pparg, Tgl (also known as Pnpla2), Plin1 (also known as Plin) and Lpl in white adipose tissue. r, s, Urinary levels of norepinephrine (r) and epinephrine (s) in Lcn2osb−/− mice and Lcn2fl/fl littermates. t, u, Indirect calorimetry measurements (t) and daily food intake normalized to body weight (u) in Lcn2osb−/− mice and Lcn2fl/fl littermates. Scale bars, 200 μm (c), 100 μm (d) and 50 μm (g). Data are mean ± s.e.m.; n = 5 (a, b, e, r–t), n = 8 (c, d, q), n = 7 (i–p), n = 3 (g) and n = 9 (u) mice per group. Results are representative of three independent experiments. *P < 0.05 (Student’s t-test).
Extended Data Figure 4 The anorexigenic function of LCN2 influences fat mass, body weight and insulin sensitivity identically in Lcn2osb−/− and Lcn2(OC)osb−/− mice.
a–f, Body weight (a), fat mass (b), insulin tolerance (c), serum insulin levels (d), glucose-stimulated insulin secretion (e) and glucose tolerance (f) in pair-fed Lcn2osb−/− and Lcn2fl/fl littermates. Data from Figs 2l, 2d, 1h, 2a, 1j and 1f, respectively, for fed Lcn2osb−/− mice are included for comparison. g, Detection of the Lcn2 floxed (primers a–b) and mutant allele, Δflox, (primers c–d) in genomic DNA isolated from tissues of Lcn2(OC)osb−/− mice. h, i, Serum levels of LCN2 (h) and tissue expression of Lcn2 (i) in Lcn2(OC)osb−/− mice and their Lcn2fl/fl littermates. j–q, glucose tolerance (j), insulin tolerance (k), glucose-stimulated insulin secretion (l), random-fed serum-insulin levels (m), fat pad mass (n), fat mass (o), food intake (p) and body weight (q) in Lcn2(OC)osb−/− mice and their Lcn2fl/fl littermates. r, Histomorphometric analysis of bone mass and Von Kossa staining of vertebral sections. BV/TV, bone volume over tissue volume; Ob.N./T.Ar, osteoblast numbers per trabecular area; OcS/BS, osteoclast surface per bone surface. s, Expression levels of osteoblastogenic and osteoclastogenic genes in bone in Lcn2osb−/− mice and their Lcn2fl/fl littermates. t, Serum osteocalcin levels in Lcn2osb−/− mice and their Lcn2fl/fl littermates. u, Lcn2 expression levels in indicated tissues following fasting–refeeding of wild-type mice. Data are mean ± s.e.m.; n = 6 (a–f, r), n = 3 (s), n = 10 (t) and n = 5 (u) mice per group; n = 3 wild-type, n = 7 Bglap-Cre, n = 7 Lcn2(OC)osb−/− and n = 6 Lcn2fl/fl mice (h–q). Results are representative of three independent experiments. *P < 0.05 (Student’s t-test).
Extended Data Figure 5 Exogenous LCN2 decreases body-weight gain and increases energy expenditure in lean and obese mice.
a–f, Wild-type mice were treated with LCN2 (150 ng g−1 per day) or vehicle for 16 weeks. a, Serum levels of LCN2. b, Body-weight gain. c, Expression levels of Cebpa and Pparg in white adipose tissue. d, Serum insulin levels. e, Pancreas insulin staining and islet histomorphometry. Scale bar, 200 μm. f, Indirect calorimetry measurements. g, h, Coomassie blue staining (g) and mass spectrometry analysis (h) of recombinant mouse LCN2. Body-weight gain (i), insulin tolerance (j) and indirect calorimetry (k) measurements of Leprdb/db mice treated with LCN2 (150 ng g−1 per day) or vehicle for 16 weeks. n = 8 mice wild-type and n = 6 Leprdb/db mice per group. Results are representative of three independent experiments. Data are mean ± s.e.m.; *P < 0.05 (a–f; Student’s t-test). #P < 0.05 (i–k) for Leprdb/db (vehicle or LCN2 treated) versus Leprdb/+ (vehicle) groups (Student’s t-test). *P < 0.05 (i–k) Leprdb/db (LCN2) versus Leprdb/+ (vehicle) and Leprdb/db (vehicle) groups (ANOVA).
Extended Data Figure 6 Increased food intake, compromised glucose metabolism and normal bone mass in Lcn2−/− mice.
a, Serum levels of leptin in Lcn2osb−/− mice and their Lcn2fl/fl littermates. b, Expression levels of leptin in white adipose tissue. c–g, Expression levels of the gastrointestinal hormones that regulate appetite Cck (c), Pyy (d), Glp1 (also known as Zglp1; e), Gip (f) and ghrelin (Ghrl; g) under fed (F), 16-h fasted (FD) and re-fed (RF) (2 h after 16-h fasting) conditions for Lcn2osb−/− mice and their Lcn2fl/fl littermates. h, Detection of the Lcn2 mutant allele, Δflox, (primers c–d) in genomic DNA isolated from tissues of Lcn2−/− mice. i, j, Serum levels of LCN2 (i) and tissue expression of Lcn2 (j) in Lcn2−/− mice and their Lcn2+/+ littermates. k–s, Analysis of Lcn2−/− mice and their Lcn2+/+ littermates. k, Glucose tolerance test. l, Insulin tolerance test. m, Glucose-stimulated insulin secretion. n, Fat pad mass. o, Fat mass. p, Food intake. q, Body weight. r, Bone histomorphometric analysis. s, Expression levels of osteoblastogenic and osteoclastogenic genes in bone. t, u, LCN2 crosses the blood–brain barrier to regulate food intake. t, LCN2 levels in the brain and serum of Lcn2−/− and wild-type littermates 2 h after i.p. administration of LCN2. u, LCN2 levels in the brain of wild-type mice 3 h after refeeding. v, LCN2 levels in the brain and serum of Lcn2−/− mice following ICV administration of 0.02 ng h−1 LCN2 or vehicle for 8 days. Data are mean ± s.e.m.; n = 10(a, b, i), n = 5 (c–g, k, l, p, r, s), n = 7 (m–o, q), n = 4 (j, t) and n = 3 (u, v) mice per group. Results are representative of three independent experiments. *P < 0.05 (Student’s t-test).
Extended Data Figure 7 MC4R, but not SLC22A17, is required for the stimulation of cAMP activity and anorexigenic gene expression induced by LCN2.
a–d, Expression levels of neuropeptides regulating appetite in the hypothalamus of Lcn2fl/fl and Lcn2osb−/− mice (a, c) and wild-type mice injected daily i.p. with LCN2 (150 ng g−1 per day) or vehicle for 16 weeks (b, d). e, o, Mc4r (e) or Slc22a17 (o) expression levels in GT1-7 hypothalamic cells following transfection with Mc4r or Slc22a17 siRNA. si-scramble, Scramble siRNA. f, g, p, cAMP production in GT1-7 cells treated with LCN2 following pretreatment with the SHU9119 inhibitor (g) or silencing of Mc4r (f) or Slc22a17 (p) expression. h, Expression levels of melanocortin receptors and LCN2 receptors in HEK293T cells transfected with an MC4R expression vector or empty vector. ND, not detected. i, j, Western blot analysis of CREB phosphorylation (i) and Fos induction (j) in GT1-7 cells treated with LCN2, vehicle or α-MSH. k–t, Expression levels of Mc4r and targets in GT1-7 cells treated with LCN2 or α-MSH for 4 h (k–n) and after silencing of Slc22a17 (q–t). u, Binding of biotinylated LCN2 and α-MSH in GT1-7 cells transiently transfected with Mc4r siRNA or scramble siRNA. v, Quantification of Fos-expressing neurons in the PVH of wild-type and Mc4r−/− mice injected with LCN2 or MT-II. Data are mean ± s.e.m. of triplicates. n = 9 (a) and n = 7 (b–d) mice per group. Results are representative of three independent experiments. *P < 0.05 (Student’s t-test).
Extended Data Figure 8 Binding affinity and activation potency of LCN2 on MC3R and MC1R.
a–f, Saturation (a, d) and competition binding (b, c, e, f) assay curves of LCN2 and α-MSH in HEK293T cells transfected with MC3R (a–c) or MC1R (d–f). Displacement of biotinylated α-MSH (b, e) or biotinylated LCN2 (c, f) by the indicated proteins. g, h, cAMP production in HEK293T cells transfected with MC3R (g) or MC1R (h) along with a cAMP/luciferase reporter and treated for 15 min as indicated. Results are representative of three independent experiments. Data are mean ± s.e.m.; i, EC50, Kd and Ki of LCN2, α-MSH and AGRP on melanocortin receptors expressed in HEK293T cells.
Extended Data Figure 9 Binding and activation of PVH neurons by LCN2.
a, Binding of biotinylated LCN2 to PVH and VMH neurons, but not neurons in the arcuate nucleus (ARC) in hypothalamic sections of Lcn2−/− and Mc4r−/− mice. Competition with unlabelled LCN2 or GST. b–d, Bright-field images of a targeted (as indicated by the arrows) neurons. b, A Sim-1 neuron from a Sim-1 cre::tdTomato mouse. c, A Pomc neuron from Pomc–hrGFP mouse. d, A Npy neuron from Npy–hrGFP mouse. The same neuron is shown in red (Alexa Fluor 594, tdTomato), green (FITC, hrGFP) and blue (Alexa Fluor 350, DAPI), as well the merged image. Scale bars, 100 μm (a) and 50 μm (b–d).
Extended Data Figure 10 MC4R is required for the improvement in glucose metabolism and energy expenditure by LCN2.
a–d, Mc4r−/− and wild-type littermates were treated with LCN2 (150 ng/g/day) or vehicle for 8 weeks. a, Indirect calorimetry measurements. b, Serum insulin levels. c, Glucose tolerance test. d, Insulin tolerance test. e–k, Serum levels of LCN2 (e), indirect calorimetry measurements (f–h), glucose tolerance test (i), insulin tolerance test (j) and glucose-stimulated insulin secretion (k) in Mc4r+/−, Lcn2osb+/−, Mc4r+/−::Lcn2osb+/− and wild-type littermates. Data are mean ± s.e.m.; n = 5 wild-type and n = 7 Mc4r−/− mice (a–d). l, LCN2 plasma levels in patients with mutated and normal MC4R sequences. n = 6 wild-type, Mc4r+/− and Mc4r+/−::Lcn2osb+/− mice and n = 5 Lcn2osb+/− mice (e–k). Results are representative of three independent experiments. *P < 0.05 indicated genotypes or treatments versus wild-type mice (Student’s t-test). #P < 0.05 when comparing either Mc4r−/− (vehicle-treated) and Mc4r−/− (LCN2-treated) groups (a) or Mc4r+/−::Lcn2osb+/− mice versus wild-type, Mc4r+/− and Lcn2osb+/− mice (f–k) (ANOVA).
Supplementary information
Supplementary Figures
The file contains gel source data, uncropped images of western, northern, southern blots and agarose gels. (PDF 3448 kb)
Supplementary Table 1
This file contains the microarray analysis of calvaria-derived osteoblasts from \({\rm Foxo1}_{{\rm osb}^{{\rm - / - }} } \) mice. Table contains genes with p<0.10, FDR<0.33 (where only 726 genes encoding for secreted proteins are taken into account in computing the FDR) and absolute value of the log2Fold change (FC)≥2, sorted by absolute value log2 FC. (XLSX 56 kb)
Source data
Rights and permissions
About this article
Cite this article
Mosialou, I., Shikhel, S., Liu, JM. et al. MC4R-dependent suppression of appetite by bone-derived lipocalin 2. Nature 543, 385–390 (2017). https://doi.org/10.1038/nature21697
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21697
This article is cited by
-
Enhancing cardiometabolic health: unveiling the synergistic effects of high-intensity interval training with spirulina supplementation on selected adipokines, insulin resistance, and anthropometric indices in obese males
Nutrition & Metabolism (2024)
-
Osteocyte-derived sclerostin impairs cognitive function during ageing and Alzheimer’s disease progression
Nature Metabolism (2024)
-
Myeloid-derived grancalcin instigates obesity-induced insulin resistance and metabolic inflammation in male mice
Nature Communications (2024)
-
Stress increases hepatic release of lipocalin 2 which contributes to anxiety-like behavior in mice
Nature Communications (2024)
-
Linea osteoblastica-osteocitaria e produzione ormonale
L'Endocrinologo (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.