Abstract
Adipose tissue is a major site of energy storage and has a role in the regulation of metabolism through the release of adipokines. Here we show that mice with an adipose-tissue-specific knockout of the microRNA (miRNA)-processing enzyme Dicer (ADicerKO), as well as humans with lipodystrophy, exhibit a substantial decrease in levels of circulating exosomal miRNAs. Transplantation of both white and brown adipose tissue—brown especially—into ADicerKO mice restores the level of numerous circulating miRNAs that are associated with an improvement in glucose tolerance and a reduction in hepatic Fgf21 mRNA and circulating FGF21. This gene regulation can be mimicked by the administration of normal, but not ADicerKO, serum exosomes. Expression of a human-specific miRNA in the brown adipose tissue of one mouse in vivo can also regulate its 3′ UTR reporter in the liver of another mouse through serum exosomal transfer. Thus, adipose tissue constitutes an important source of circulating exosomal miRNAs, which can regulate gene expression in distant tissues and thereby serve as a previously undescribed form of adipokine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Krol, J., Loedige, I. & Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 (2010)
Sun, L. et al. Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 13, 958–965 (2011)
Trajkovski, M. et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653 (2011)
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009)
Ameres, S. L. & Zamore, P. D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475–488 (2013)
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108, 5003–5008 (2011)
Thery, C., Amigorena, S., Raposo, G. & Clayton, A. Curr Protoc Cell Biol Chapter 3, Unit 3 22 (2006)
György, B. et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 68, 2667–2688 (2011)
Hata, A. & Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 8, re3 (2015)
Dumortier, O., Hinault, C. & Van Obberghen, E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 18, 312–324 (2013)
Arner, E. et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 61, 1986–1993 (2012)
Capobianco, V. et al. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J. Proteome Res. 11, 3358–3369 (2012)
Caroli, A., Cardillo, M. T., Galea, R. & Biasucci, L. M. Potential therapeutic role of microRNAs in ischemic heart disease. J. Cardiol. 61, 315–320 (2013)
Guay, C., Roggli, E., Nesca, V., Jacovetti, C. & Regazzi, R. Diabetes mellitus, a microRNA-related disease? Transl. Res. 157, 253–264 (2011)
Mori, M. A. et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 16, 336–347 (2012)
Mori, M. A. et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J. Clin. Invest. 124, 3339–3351 (2014)
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008)
Taylor, D. D., Zacharias, W. & Gercel-Taylor, C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 728, 235–246 (2011)
Escola, J. M. et al. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 (1998)
Février, B. & Raposo, G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16, 415–421 (2004)
Ortega, F. J. et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One 5, e9022 (2010)
Chou, W. W. et al. Decreased microRNA-221 is associated with high levels of TNF-α in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell. Physiol. Biochem. 32, 127–137 (2013)
Oger, F. et al. Cell-specific dysregulation of microRNA expression in obese white adipose tissue. J. Clin. Endocrinol. Metab. 99, 2821–2833 (2014)
Keller, P. et al. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 11, 7 (2011)
McGregor, R. A. & Choi, M. S. microRNAs in the regulation of adipogenesis and obesity. Curr. Mol. Med. 11, 304–316 (2011)
Potthoff, M. J., Kliewer, S. A. & Mangelsdorf, D. J. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 26, 312–324 (2012)
Badman, M. K. et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 5, 426–437 (2007)
Wong, N. & Wang, X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146–D152 (2015)
Yao, Y. et al. MicroRNA profiling of human gastric cancer. Mol. Med. Rep. 2, 963–970 (2009)
Uhrig-Schmidt, S. et al. Gene delivery to adipose tissue using transcriptionally targeted rAAV8 vectors. PLoS One 9, e116288 (2014)
Atai, N. A. et al. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J. Neurooncol. 115, 343–351 (2013)
Zech, D., Rana, S., Büchler, M. W. & Zöller, M. Tumor-exosomes and leukocyte activation: an ambivalent crosstalk. Cell Commun. Signal. 10, 37 (2012)
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007)
Blüher, M. Adipokines - removing road blocks to obesity and diabetes therapy. Mol. Metab. 3, 230–240 (2014)
Théry, C., Ostrowski, M. & Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009)
Bang, C. et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 124, 2136–2146 (2014)
Hergenreider, E. et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 14, 249–256 (2012)
Mittelbrunn, M. et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282 (2011)
Ismail, N. et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121, 984–995 (2013)
Zernecke, A. et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2, ra81 (2009)
van der Vos, K. E. et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro-oncol. 18, 58–69 (2016)
Yuan, A. et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One 4, e4722 (2009)
Gallo, A., Tandon, M., Alevizos, I. & Illei, G. G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 7, e30679 (2012)
Turchinovich, A., Weiz, L., Langheinz, A. & Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011)
Tian, Y. et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 35, 2383–2390 (2014)
Mathivanan, S., Fahner, C. J., Reid, G. E. & Simpson, R. J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, D1241–D1244 (2012)
Mestdagh, P. et al. A novel and universal method for microRNA RT–qPCR data normalization. Genome Biol. 10, R64 (2009)
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004)
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998)
Tran, T. T., Yamamoto, Y., Gesta, S. & Kahn, C. R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 7, 410–420 (2008)
Acknowledgements
We thank M. Torriani and K. V. Fitch for assistance with HIV lipodystrophy samples; M. Lynnes, S. Kasif, and A. M. Cypess for help with reagents and discussions; and the Joslin Histology, Media and Physiology Core Facilities for help with experiments. This study was supported by grants from the NIH R01 DK082659 and R01 DK033201, the Mary K. Iacocca Professorship, and the Joslin Diabetes Center DRC Grant P30DK036836. S.K.G. was funded by grants from the NIH (P30 DK040561). M.A.M. was funded by grants from FAPESP (2010/52557-0 and 2015/01316-7).
Author information
Authors and Affiliations
Contributions
M.A.M. assisted with experimental design, generated the ADicerKO mice and designed the Ad-Luc-FGF2-3′-UTR constructs; J.M.D. carried out bioinformatics analysis; M.K. performed adenoviral injections in BAT; M.S. assisted with retro-orbital injections; C.W. created Ad-lacZ, Ad-pre-hsa-miR302f and Ad-Luc-miR302f-3′-UTR adenoviruses; T.N.R. assisted with retro-orbital and tail vain injections; J.N.W. assisted with fat depot miRNA PCR; R.G.-M. assisted with IVIS experiments and in vitro luminescence assays; S.K.G. provided human HIV lipodystrophy serum samples; P.G. provided human CGL sera samples; and T.T. and C.R.K. designed the study, collected and analysed data, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
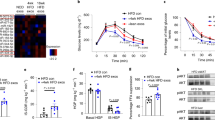
Extended Data Figure 1 Fat is a major source of circulating exosomal miRNAs in mice.
a, Electron micrograph of exosomes isolated from ADicerKO serum by differential centrifugation. b, EXOCET ELISA assay measuring CETP protein in exosome samples, corresponding to isolated exosome number from serum of ADicerKO (KO) or Lox (WT) mice. c, qNano assay measuring the number and size of exosomes, based on tunable resistive pulse sensing technology from exosome preparations from ADicerKO (top) or Lox (bottom) mice. d, Principle component analysis of exosomal miRNA levels in ADicerKO and Lox mice, n = 4 per group. Data are mean s.e.m.
Extended Data Figure 2 Adipose tissue Dicer regulates exosomal and serum miRNA content.
a, Heatmap showing Z scores of miRNA expression in whole serum from ADicerKO (KO) and wild-type mice (WT), and exosomal miRNAs from ADicerKO (ExoKO) or wild-type mice (ExoWT) (n = 4 per group). b, Heatmap showing Z scores of expression of exosomal miRNAs from culture supernatant of Dicerfl/fl pre-adipocytes transfected with adenovirus encoding GFP or Cre (n = 3 per group). c, Heatmap showing Z scores of miRNA expression measurements of exosomal miRNAs from the serum of 4-week-old ADicerKO (AdicerKO) and Lox (Control) mice (n = 3 per group).
Extended Data Figure 3 Fat is a major source of circulating exosomal miRNAs in humans.
a, Demographic information about human patients with HIV-associated lipodystrophy, CGL (labelled as Lipodystrophy) or control subjects. b, EXOCET ELISA assay measuring CETP protein as a measure of exosome number from isolated from human sera of individuals with HIV-associated lipodystrophy (HIV), CGL and control subjects (n = 4 per group). c, Principle component analysis of exosomal miRNA expression in patients with HIV-associated lipodystrophy or CGL, or in control subjects (n = 4 per group). Data are mean ± s.e.m.
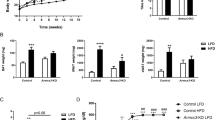
Extended Data Figure 4 Transplant donor fat depot miRNA signatures are distinct.
a, Principle component analysis of miRNA expression in mouse epididymal (EPI), inguinal (ING) and BAT fat depots (n = 4 per group). b, Weight of the transplanted epididymal WAT, inguinal WAT and BAT at time of transplantation into ADicerKO mice (white bars) and at time of death (chequered bars) (n = 3). c, Weight of ADicerKO mice that underwent sham surgery (Sal) or transplantation with epididymal, inguinal, or BAT fat; Lox mice that underwent sham surgery (WT) serve as a control. Data are mean ± s.e.m.
Extended Data Figure 5 Fat tissue transplantation alters exosomal miRNA content.
a, Principle component analysis of serum exosomal miRNA levels in ADicerKO mice after sham surgery (Sal) or transplantation with inguinal fat (ING), epididymal fat (EPI) or BAT; Lox littermates that underwent sham surgery serve as a control (n = 4 per group). b, Circulating levels of insulin and the adipokines IL-6, leptin and adiponectin in the groups of mice in a. Two-tailed t-test, P < 0.05 (n = 3 per group).
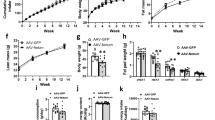
Extended Data Figure 6 ADicerKO mice exhibit reduced Fgf21 abundance.
a, Levels of Fgf21 mRNA, as assessed by qRT–PCR in liver, BAT, inguinal (ING), epididymal (Epi), pancreas (Panc), kidney (Kidn) and quadriceps muscle (Quad) tissue from ADicerKO mice (black bars) or Lox littermates (white bars). P = 0.0286 by two-tailed Mann–Whitney U test (n = 4 per group). b, Relative abundance (shown as log2 fold change (log2FC)) as assessed by qRT–PCR of miR-99a, miR-99b, and miR-100 in exosomes extracted from ADicerKO (KO) mice that underwent sham or fat-transplantation surgery (as in Fig. 2e) and wild-type mice that underwent sham surgery (n = 4 per group). Data are mean ± s.e.m. *P = 0.05.
Extended Data Figure 7 Fgf21 is regulated by exosomal fat-derived miRNAs in vitro.
a, Fgf21 3′ UTR luciferase-reporter activity in AML-12 mouse liver cells after transfection with 10 nM miR-99a, miR-99b, miR-100, miR-466i or control by direct electroporation. P = 0.003 by two-tailed t-test (n = 3 per group). b, Abundance of Fgf21 mRNA in AML-12 mouse liver cells following transfection with 10 nM miRNA miR-99a, miR-99b, miR-100 or miR-466i. P = 0.037, two tailed t-test (n = 3 per group). c, Hepatic Fgf21 mRNA levels by qRT–PCR followed by a 48-h incubation of AML-12 hepatic cells with exosomes derived from Lox or ADicerKO (−) mice, or with exosomes from ADicerKO mice electroporated with 10 nM miR-99a, miR-99b, miR-100 or miR-466i. P = 0.0001, two-tailed t-test (n = 3 per group). Data are mean ± s.e.m. *P = 0.05.
Extended Data Figure 8 Adipose tissue-specific miRNAs are enriched in the liver after fat transplantation.
a, qRT–PCR quantification of levels of mature miR-16, miR-201, and miR-222 in liver from Lox mice, ADicerKO mice and ADicerKO mice transplanted with BAT (KO + BAT). P = 0.02 for miR-16, P = 0.002 for miR-201, and P = 0.028 for miR-222; one-way ANOVA; significant comparisons were identified by Tukey’s multiple comparisons test (n = 3 per group). b, qRT–PCR quantification of the levels of pre-miR-16, pre-miR-201, and pre-miR-222 in the livers of Lox mice, ADicerKO mice and ADicerKO mice transplanted with BAT. P < 0.05, one-way ANOVA (n = 3 per group,). c, qRT–PCR-derived Ct values of adenoviral DNA isolated from BAT and liver tissue in Protocol 1 (BAT-p1 and Liver-p1, respectively), and from liver tissue in Protocol 2 (liver-p2), detecting adenoviral lacZ or pre-miR-302f (n = 4 per group). Data are mean ± s.e.m. *P = 0.05.
Supplementary information
Supplementary Table 1
qPCR analysis of exosomal miRNA from sera of 6 month old male ADicerKO mice vs. controls. (CSV 38 kb)
Supplementary Table 2
qPCR analysis of exosomal miRNA from sera of human HIV lipodystrophy subjects vs. controls (CSV 32 kb)
Supplementary Table 3
qPCR analysis of exosomal miRNA from sera of human CGL subjects vs. controls (CSV 32 kb)
Supplemental Table 4
miRNA directions of change (1: increase; -1: decrease; 0: non-significant) in lipodystrophy subjects vs controls corresponding to Venn diagram in Figure 1g (CSV 5 kb)
Supplemental Table 5
List of serum exosomal miRNAs that are down-regulated in both human lipodystrophies and ADicerKO mice (CSV 0 kb)
Supplemental Table 6
qPCR analysis of mouse fat depots vs. controls (CSV 160 kb)
Supplemental Table 7
Logical values indicating whether transplantation of the fat depot could reconstitute the miRNA (TRUE), or not (FALSE); these values correspond to the Venn diagram in Figure 2c (CSV 5 kb)
Supplemental Table 8
qPCR analysis of exosomal miRNA from serum of mouse after fat transplants (or WT) vs. saline knockout (SAL) (CSV 49 kb)
Source data
Rights and permissions
About this article
Cite this article
Thomou, T., Mori, M., Dreyfuss, J. et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 (2017). https://doi.org/10.1038/nature21365
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21365
This article is cited by
-
Extracellular vesicle therapy for obesity-induced NAFLD: a comprehensive review of current evidence
Cell Communication and Signaling (2024)
-
Schwann cell-derived extracellular vesicles promote memory impairment associated with chronic neuropathic pain
Journal of Neuroinflammation (2024)
-
Neonatal adiposity is associated with microRNAs in adipocyte-derived extracellular vesicles in maternal and cord blood, a discovery analysis
International Journal of Obesity (2024)
-
Liver-derived extracellular vesicles improve whole-body glycaemic control via inter-organ communication
Nature Metabolism (2024)
-
miR-548ag promotes DPP4 expression in hepatocytes through activation of TLR(7/8)/NF-κB pathway
International Journal of Obesity (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.