Abstract
The maternal-to-zygotic transition (MZT) is one of the most profound and tightly orchestrated processes during the early life of embryos, yet factors that shape the temporal pattern of vertebrate MZT are largely unknown. Here we show that over one-third of zebrafish maternal messenger RNAs (mRNAs) can be N6-methyladenosine (m6A) modified, and the clearance of these maternal mRNAs is facilitated by an m6A-binding protein, Ythdf2. Removal of Ythdf2 in zebrafish embryos decelerates the decay of m6A-modified maternal mRNAs and impedes zygotic genome activation. These embryos fail to initiate timely MZT, undergo cell-cycle pause, and remain developmentally delayed throughout larval life. Our study reveals m6A-dependent RNA decay as a previously unidentified maternally driven mechanism that regulates maternal mRNA clearance during zebrafish MZT, highlighting the critical role of m6A mRNA methylation in transcriptome switching and animal development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Tadros, W. & Lipshitz, H. D. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 (2009)
Lee, M. T., Bonneau, A. R. & Giraldez, A. J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613 (2014)
Walser, C. B. & Lipshitz, H. D. Transcript clearance during the maternal-to-zygotic transition. Curr. Opin. Genet. Dev. 21, 431–443 (2011)
Giraldez, A. J. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 (2006)
Mishima, Y. & Tomari, Y. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol. Cell 61, 874–885 (2016)
Mathavan, S. et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 1, 260–276 (2005)
Aanes, H. et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 21, 1328–1338 (2011)
Rabani, M. et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 159, 1698–1710 (2014)
Desrosiers, R., Friderici, K. & Rottman, F. Characterization of Novikoff Hepatoma messenger-RNA methylation. Fed. Proc. 34, 628–628 (1975)
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011)
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013)
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014)
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014)
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 (2015)
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014)
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014)
Kane, D. A. & Kimmel, C. B. The zebrafish midblastula transition. Development 119, 447–456 (1993)
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012)
Chen, K. et al. High-resolution N6 -methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int. Ed. 54, 1587–1590 (2015)
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015)
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012)
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013)
Gerber, A. P., Luschnig, S., Krasnow, M. A., Brown, P. O. & Herschlag, D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 4487–4492 (2006)
Laver, J. D. et al. Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition. Genome Biol. 16, 94 (2015)
Stoeckius, M. et al. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. EMBO J. 33, 1751–1766 (2014)
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995)
Thisse, B. & Thisse, C. In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol. Biol. 1211, 53–67 (2014)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010)
de Hoon, M. J. L., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004)
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 (2009)
Huang, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009)
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011)
Bazzini, A. A., Lee, M. T. & Giraldez, A. J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 (2012)
Lee, M. T. et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013)
Acknowledgements
This work was supported by National Institutes of Health HG008688, GM113194 (both to C.H.), and HD072598 (to R.K.H.). C.H. is an investigator of the Howard Hughes Medical Institute (HHMI). B.S.Z. is an HHMI International Student Research fellow. The Mass Spectrometry Facility of the University of Chicago is funded by National Science Foundation (CHE-1048528). We thank H. Pickersgill for editing help.
Author information
Authors and Affiliations
Contributions
B.S.Z., X.W. and A.V.B. contributed equally to this work. B.S.Z., X.W., A.V.B., C.H. and R.K.H. designed experiments. B.S.Z., X.W., A.V.B. and H.S. performed biochemistry experiments. A.V.B., B.S.Z., X.W. and A.K. performed cell biology experiments. B.S.Z., X.W. and Z.L. analysed sequencing data. B.S.Z., X.W., A.V.B. and C.H. wrote the manuscript with inputs from Z.L. and R.K.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks E. Woon and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Generation and characterization of ythdf2 mutant fish.
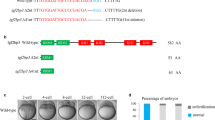
a, Design of transcription activator-like effectors (TALEs) used to generate ythdf2 mutant fish. An 8 bp deletion upstream of the YTH RNA-binding domain is made to create a premature stop codon. b, Photos of adult wild-type and ythdf2−/− mutant fish. Homozygous ythdf2 mutant males are often smaller than wild-type males. c, Table showing the percentage of embryos suffering from MBT-period developmental delay and lethality for crosses between different homozygous genotypes. Homozygous mutant females produce ~95% embryos that suffer from MBT-period developmental delay, regardless of paternal genotype. Homozygous mutant males produce on average ~70% embryos that do not develop past the one-cell stage, regardless of maternal genotype. These two phenotypes can segregate independently from one another and reveal that Ythdf2 is pleiotropic in zebrafish development (see Supplementary Discussion). Each row is tabulated from 6 to 18 crosses, scoring between 365 and 1,026 embryos, and using randomly selected males and females from each genotype from at least 4 fish of each sex. d, Table showing additional crosses to characterize the paternal-associated lethality and/or sterility of ythdf2 loss of function. Heterozygous males produce about half the number one-cell arrested offspring as homozygous mutant males, perhaps indicating a function for Ythdf2 in later gametogenesis; this arrest occurs without respect to maternal genotype. Data for each row is tabulated from 5 crosses each, using randomly selected males and females from each genotype from at least 4 fish of each sex.
Extended Data Figure 2 Phenotypes of maternal ythdf2−/− mutant embryos.
a, Still images taken from the time-lapse movie of a maternal ythdf2−/− mutant embryo filmed between 2.25 h.p.f. and 6.55 h.p.f. highlighting the development delay phenotype occurring during the MBT period. Red texts indicate stages experiencing the delay. All cell divisions are normal in mutant embryos until the cell cycle that should take place between high and oblong stages, cell cycle 12. The high stage in mutant embryos lasts ~60 min while the similar stage in wild-type lasts ~20 min. Maternal ythdf2−/− mutants lose another ~30 min of developmental time between the pseudo-dome and pseudo-50% epiboly stages before reaching the 50% epiboly stage at 6.5 h.p.f. versus 5 h.p.f. in wild-type. b, Injection of ythdf2 mRNA allows partial rescue of the ythdf2 mutant phenotype. Maternal ythdf2−/− mutant embryos injected with 150 pg ythdf2 mRNA showed an overall reduction in the cell cycle 12 delay compared to uninjected embryos of the same genotype. In these partially rescued embryos, an appreciable number of cells during cell cycle 12 divided at the same time as wild-type cells did. This partial rescue is nearly 100% penetrant (n = 69, four experiments). c, Immunofluorescent microscopy of wild-type, ythdf2 mutant, and ythdf2 mRNA-injected mutant embryos fixed at 3 h 50 min post-fertilization and stained with anti-pH3 antibody to label nuclei in late G2 phase through M-phase of the cell cycle. The bar chart shows the average number of condensed pH3-positive nuclei per 100 cells. The results showed that unlike wild-type embryos, maternal ythdf2−/− mutants are lacking in the tight, condensed pH3 nuclei staining characteristic of metaphase/anaphase, which can be partially rescued by the injection of ythdf2 mRNA. Error bars, mean ± s.e.m., n = 8 embryos counted for each condition, 100 cells each embryo. P values were determined using unpaired Student’s t-test. Scale bar, 200 μm.
Extended Data Figure 3 Characterization of m6A modification and gene expression change in the early embryonic transcriptome of zebrafish.
a, Venn diagrams show the overlap of two replicates of m6A-modified transcripts determined by m6A-seq and m6A-CLIP-seq at five time points. b, Distribution of m6A-sites (number of m6A-seq peaks) in the six gene groups over time. c, Number of m6A-modified genes identified in both m6A-seq and m6A-CLIP-seq at five time points. d, Quantification of the m6A/(G+A+C+U) ratio of mRNAs purified from zebrafish embryos at various developmental time points by LC-MS/MS. The fluctuation observed may reflect the complex dynamics of the decay of the methylated maternal transcripts, the transcription and methylation of newly synthesized zygotic mRNAs, and their subsequent decay during early development. All time points were normalized to 0 h.p.f. value. Error bars, mean ± s.d., n = 3 (technical replicates). e, Consensus motif identified by HOMER with m6A-CLIP peaks at five time points. f, Metagene profiles depict the subtranscript distribution pattern of m6A-sites (from m6A-seq) within the zebrafish transcriptome. m6A-seq peak signals are enriched after the start codon and before the stop codon. g, h, Gene Ontology (GO) analysis of non-methylated maternal transcripts (g) and methylated ones (h). i, Table showing the number of genes downregulated and upregulated in maternal ythdf2−/− mutants at 4 h.p.f. within the 6 gene expression clusters (top 20%).
Extended Data Figure 4 Design and knockdown efficiency of the morpholino targeting ythdf2.
a, Design of ythdf2 morpholino that targets the intersection of the 5′ UTR and CDS of ythdf2 mRNA. b, Efficiency test of ythdf2 morpholino treatment inhibiting the expression of Ythdf2 by western blot. mRNAs containing the 5′ UTR and full-length CDS of ythdf2 with Flag-tag sequence fused at the C terminus were co-injected with either control or ythdf2 MO into single-cell wild-type embryos. Ythdf2–Flag was detected by the anti-Flag antibody. Material from six embryos was loaded into each lane. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 5 Overlap between methylated genes and upregulated genes in maternal ythdf2−/− embryos.
a, Quantification of the m6A/A ratio of the total mRNA purified from maternal ythdf2−/− and wild-type embryos by LC-MS/MS. P values were determined using two-sided Student’s t-test for paired samples. Error bars, mean ± s.d. of two technical replicates (data points with same colour) from two biological experiments. b, Composition of upregulated, methylated genes in maternal ythdf2−/− samples at 4 h.p.f. A total of 3,292 genes are both methylated and upregulated upon ythdf2 loss of function, over 60% of which are maternal genes. c, Venn diagram depicts the overlapping of m6A-modified transcripts determined by m6A-seq, m6A-CLIP-seq, and differentially expressed transcripts between ythdf2 loss of function samples and controls at 4 h.p.f. The 135 intersected genes were defined as the most stringent Ythdf2 RNA targets. Without including the MO-treated result (but with ythdf2−/− result), we can estimate about 383 Ythdf2 target mRNAs. d, GO analysis of the most stringent Ythdf2 target genes.
Extended Data Figure 6 Methylation profiles of Ythdf2 target transcripts.
a, Examples of Ythdf2 RNA targets harbouring both m6A-seq peaks and m6A-CLIP-seq peaks. Coverage of m6A immunoprecipitation (IP) and input fragments are indicated in red and blue, respectively. m6A-CLIP-seq peaks are highlighted in green. Grey lines signify CDS borders. b, The dynamics of m6A immunoprecipitation signals over time in Ythdf2 RNA targets. Coverage of m6A immunoprecipitation and input fragments are indicated in red and grey, respectively. The locations of consensus motifs of GGACU and AGACU are indicated as blue dots. Red lines show splice junctions. Blue squares mark the peaks identified from the m6A-seq signals. Example genes shown: buc, mtus1a, mylipa, setdb1a, tdrd1, vps26a, and zgc:162879.
Extended Data Figure 7 Degradation of reporter mRNAs in ythdf2 loss of function and control embryos.
The in vivo degradation of GFP-m6A mRNAs was faster than GFP-A mRNAs when co-injected with control MO. By contrast, GFP-m6A mRNAs displayed an undistinguishable decay rate from GFP-A mRNAs when co-injected with ythdf2 MO. The abundances of GFP-m6A and GFP-A mRNAs were determined by RT–qPCR. Error bars, mean ± s.d., n = 3 (technical replicates). P values were determined using two-sided Student’s t-test for two samples with equal variance.
Extended Data Figure 8 Zygotic ythdf2 facilitates continued regulation of maternal mRNA clearance during late MZT (6–8 h.p.f.).
a–d, Cumulative distribution of the log2 fold changes of RNA expression after 4 h.p.f. for the six gene groups between maternal ythdf2−/− (m-KO) and wild-type (a, b), or maternal–zygotic ythdf2−/− (mz-KO) and wild-type (c, d). Colour code and gene numbers of each gene group are listed under each figure panel. Compared to the relatively unchanged curves of semi-stable gene groups (black and grey), the right shift of blue curves indicates the increase of maternal RNAs while the left shift of red curves indicates the decrease of zygotic RNAs in mutant samples versus wild-type ones. Whereas the gene expression patterns of m-KO samples remain largely unchanged at 6 h.p.f. (a) and 8 h.p.f. (b), mz-KO samples show pronounced retention of maternal genes and delayed activation of zygotic genes at 6 h.p.f. (c) and continued notable delay of zygotic expression at 8 h.p.f. (d), indicating zygotically expressed Ythdf2 also functions on maternal mRNA during late MZT. Gene group categorization was based on the expression pattern data from 0–8 h.p.f. P values were calculated using two-sided Kruskal–Wallis test. Representative data shown from two independent experiments.
Extended Data Figure 9 Ythdf2 does not cause transcriptome-wide zygotic gene expression change during the zygotic stage of zebrafish early development (12–48 h.p.f.).
Cumulative distribution of the log2 fold changes of RNA expression during the zygotic stage of zebrafish early development for the six gene groups between mutant and wild-type embryos. a, b, mz-KO samples exhibit largely unchanged gene expression patterns at 12 h.p.f. (a) and 24 h.p.f. (b) versus wild-type, indicating that Ythdf2 does not affect the degradation of zygotic mRNA on the organismal level during this time period. c, Considering the potential time-matched inaccuracy due to the delayed zygotic gene expression in the mutant embryos, we performed a stage-matched comparison at the 14–17 somite stage (wild type 16 h.p.f. versus knockout 17.5 h.p.f.) and observed very similar, unchanged gene expression profiles between mutant and wild-type embryos. Both time-matched and stage-matched comparisons reveal largely indistinguishable zygotic transcriptome profiles during the investigated zygotic stage corresponds nicely with the lack of abnormal developmental phenotypes in ythdf2 mz-KO embryos during this time period. d–f, To avoid the involvement of paternal effect from male ythdf2 KO fish, we used maternal KO embryos (from wild-type male) injected with ythdf2-MO (m-KO + MO) to achieve maternal and zygotic depletion of Ythdf2. We investigated the RNA expression of all genes between wild-type and ythdf2 loss of function groups during later developmental stages and observed largely unchanged gene expression patterns in m-KO and MO samples at 24 h.p.f. (d), 36 h.p.f. (e), and 48 h.p.f. (f) versus wild-type, which indicate that Ythdf2 does not affect the degradation of zygotic mRNA on the transcriptome level during late zygotic stages. Gene group categorization was based on the expression pattern data from 0–8 h.p.f. P values were calculated using two-sided Kruskal–Wallis test. Representative data shown from two independent experiments.
Extended Data Figure 10 Target overlap of Ythdf2 and miR-430 regulation pathways.
a, Target overlap between Ythdf2 and miR-430. Genes with more than 1.5-fold upregulation upon removal of maternal Ythdf2 or miR-430 (ref. 4) were used as corresponding targets in the analysis. b, GO analysis of the common target genes of Ythdf2 and miR-430. c–f, Differential temporal regulation of the targets of miR430 and Ythdf2. In wild-type embryos, common targets are decayed first, followed by Ythdf2-specific targets, and then miR430-specific targets (c). Upon maternal ythdf2 knockout, the decay of the miR430-specific targets showed minimal delay (d), whereas the Ythdf2-specific targets and the common targets show more noticeable decay delays; the latter finding is consistent with Ythdf2 as a maternally supplied factor able to degrade maternal mRNAs earlier in time (e, f). Relative expression level is calculated by the median of the expression levels of all genes and normalized to the value at 0 h.p.f., respectively. g, Maternal ythdf2−/− embryos show a similar zygotic genome activation delay as Nanog, Sox19b, and Pou5f3 loss-of-function embryos at 4 h.p.f. The log2 fold change of the mRNA-seq data of maternal ythdf2−/− mutants and the published mRNA-seq data with combined loss of nanog, sox19b (soxB1), and pou5f3 (oct4 homologue) were plotted in the same diagram. Zygotic genes (red dots) were separately plotted out from all genes (grey dots). Intersections of crosslines mark the median points of each group of genes. h, Proposed mechanism of Ythdf2 regulating zebrafish MZT: Ythdf2-mediated RNA decay ensures the timely clearance of maternal RNA; without Ythdf2, m6A-modified maternal RNA are overrepresented in the transcriptome during MZT, which causes delayed zygotic genome activation and a substantial developmental delay.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Figure 1 showing the uncropped gel image for Extended Data Figure 4b and legends for Supplementary Data files 1 and 2 (see separate excel files). (PDF 163 kb)
Supplementary Data 1
This file contains Supplementary Data 1 – see page 7 of the Supplementary Information file for more details. (XLSX 47 kb)
Supplementary Data 2
This file contains Supplementary Data 2 – see page 7 of the Supplementary Information file for more details. (XLSX 4390 kb)
Rights and permissions
About this article
Cite this article
Zhao, B., Wang, X., Beadell, A. et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017). https://doi.org/10.1038/nature21355
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21355
This article is cited by
-
miR-430 regulates zygotic mRNA during zebrafish embryogenesis
Genome Biology (2024)
-
Cell-type-specific mRNA transcription and degradation kinetics in zebrafish embryogenesis from metabolically labeled single-cell RNA-seq
Nature Communications (2024)
-
Emerging role of RNA modification and long noncoding RNA interaction in cancer
Cancer Gene Therapy (2024)
-
Mycobacterium tuberculosis inhibits METTL14-mediated m6A methylation of Nox2 mRNA and suppresses anti-TB immunity
Cell Discovery (2024)
-
The antagonistic effect of FTO on METTL14 promotes AKT3 m6A demethylation and the progression of esophageal cancer
Journal of Cancer Research and Clinical Oncology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.