Abstract
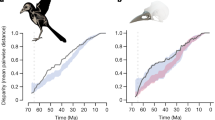
The origin and expansion of biological diversity is regulated by both developmental trajectories1,2 and limits on available ecological niches3,4,5,6,7. As lineages diversify, an early and often rapid phase of species and trait proliferation gives way to evolutionary slow-downs as new species pack into ever more densely occupied regions of ecological niche space6,8. Small clades such as Darwin’s finches demonstrate that natural selection is the driving force of adaptive radiations, but how microevolutionary processes scale up to shape the expansion of phenotypic diversity over much longer evolutionary timescales is unclear9. Here we address this problem on a global scale by analysing a crowdsourced dataset of three-dimensional scanned bill morphology from more than 2,000 species. We find that bill diversity expanded early in extant avian evolutionary history, before transitioning to a phase dominated by packing of morphological space. However, this early phenotypic diversification is decoupled from temporal variation in evolutionary rate: rates of bill evolution vary among lineages but are comparatively stable through time. We find that rare, but major, discontinuities in phenotype emerge from rapid increases in rate along single branches, sometimes leading to depauperate clades with unusual bill morphologies. Despite these jumps between groups, the major axes of within-group bill-shape evolution are remarkably consistent across birds. We reveal that macroevolutionary processes underlying global-scale adaptive radiations support Darwinian9 and Simpsonian4 ideas of microevolution within adaptive zones and accelerated evolution between distinct adaptive peaks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
30 November 2017
Please see accompanying Corrigendum (http://doi.org/10.1038/nature24665). Two missing Supplementary Information files (‘Prum_merge_taxonomy_CRC_v2.csv’ and ‘PrumTreeMerge_CRC_v1.csv’) have been added to this Letter.
References
Bright, J. A., Marugán-Lobón, J., Cobb, S. N. & Rayfield, E. J. The shapes of bird beaks are highly controlled by nondietary factors. Proc. Natl Acad. Sci. USA 113, 5352–5357 (2016)
Lamichhaney, S. et al. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375 (2015)
Phillimore, A. B. & Price, T. D. Density-dependent cladogenesis in birds. PLoS Biol. 6, e71 (2008)
Simpson, G. G. Tempo and mode in evolution. (Columbia University Press, 1944)
Ezard, T. H. & Purvis, A. Environmental changes define ecological limits to species richness and reveal the mode of macroevolutionary competition. Ecol. Lett. 19, 899–906 (2016)
Price, T. Speciation in Birds. 1st edn (Roberts and Co., 2008)
Price, T. D. et al. Niche filling slows the diversification of Himalayan songbirds. Nature 509, 222–225 (2014)
Losos, J. B. & Mahler, D. L. in Evolution since Darwin: The First 150 Years (eds M. A. Bell, D. J. Futuyma, W. F. Eanes & J. S. Levinton ) 381–420 (Sinauer Associates, 2010)
Reznick, D. N. & Ricklefs, R. E. Darwin’s bridge between microevolution and macroevolution. Nature 457, 837–842 (2009)
Alfaro, M. E. et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13410–13414 (2009)
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012)
Hedges, S. B., Marin, J., Suleski, M., Paymer, M. & Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015)
Venditti, C., Meade, A. & Pagel, M. Multiple routes to mammalian diversity. Nature 479, 393–396 (2011)
Etienne, R. S. et al. Diversity-dependence brings molecular phylogenies closer to agreement with the fossil record. Proc. Biol. Sci. 279, 1300–1309 (2012)
Rabosky, D. L. & Glor, R. E. Equilibrium speciation dynamics in a model adaptive radiation of island lizards. Proc. Natl Acad. Sci. USA 107, 22178–22183 (2010)
Jønsson, K. A. et al. Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc. Natl Acad. Sci. USA 109, 6620–6625 (2012)
Rabosky, D. L. et al. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4, 1958 (2013)
Grant, P. R. Ecology and Evolution of Darwin’s finches. 2nd edn (Princeton University Press, 1999)
Pigot, A. L., Trisos, C. H. & Tobias, J. A. Functional traits reveal the expansion and packing of ecological niche space underlying an elevational diversity gradient in passerine birds. Proc. Biol. Sci. 283, 20152013 (2016)
Lovette, I. J., Bermingham, E. & Ricklefs, R. E. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc. Biol. Sci. 269, 37–42 (2002)
Kraft, N. J. B., Valencia, R. & Ackerly, D. D. Functional traits and niche-based tree community assembly in an Amazonian forest. Science 322, 580–582 (2008)
Mitchell, J. S. Extant-only comparative methods fail to recover the disparity preserved in the bird fossil record. Evolution 69, 2414–2424 (2015)
Finarelli, J. A. & Goswami, A. Potential pitfalls of reconstructing deep time evolutionary history with only extant data, a case study using the canidae (mammalia, carnivora). Evolution 67, 3678–3685 (2013)
Slater, G. J. Iterative adaptive radiations of fossil canids show no evidence for diversity-dependent trait evolution. Proc. Natl Acad. Sci. USA 112, 4897–4902 (2015)
Ricklefs, R. E. Small clades at the periphery of passerine morphological space. Am. Nat. 165, 651–659 (2005)
Robinson, M. R. & Beckerman, A. P. Quantifying multivariate plasticity: genetic variation in resource acquisition drives plasticity in resource allocation to components of life history. Ecol. Lett. 16, 281–290 (2013)
Hadfield, J. D. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22 (2010)
Hughes, M., Gerber, S. & Wills, M. A. Clades reach highest morphological disparity early in their evolution. Proc. Natl Acad. Sci. USA 110, 13875–13879 (2013)
Mitchell, J. S. & Makovicky, P. J. Low ecological disparity in Early Cretaceous birds. Proc. Biol. Sci. 281 http://dx.doi.org/10.1098/rspb.2014.0608 (2014)
Brusatte, S. L., O’Connor, J. K. & Jarvis, E. D. The Origin and Diversification of Birds. Curr. Biol. 25, R888–R898 (2015)
Klingenberg, C. P. Visualizations in geometric morphometrics: how to read and how to make graphs showing shape changes. Hystrix 24, 15–24 (2013)
Zelditch, M. Geometric Morphometrics for Biologists: a Primer. (Elsevier Academic Press, 2004)
Fruciano, C. Measurement error in geometric morphometrics. Dev. Genes Evol. 226, 139–158 (2016)
Adams, D. C. & Otarola-Castillo, E. geomorph: an R package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399 (2013)
Gunz, P., Mitteroecker, P. & Bookstein, F. L. in Modern Morphometrics in Physical Anthropology (ed. D. E. Slide ) 73–98 (Kluwer Academic/Plenum Publishers, 2004)
Rohlf, F. J. The tps series of software. Hystrix, the Italian Journal of Mammalogy 26, 9–12 (2015)
Revell, L. J. Size-correction and principal components for interspecific comparative studies. Evolution 63, 3258–3268 (2009)
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012)
Uyeda, J. C., Caetano, D. S. & Pennell, M. W. Comparative analysis of principal components can be misleading. Syst. Biol. 64, 677–689 (2015)
Hackett, S. J. et al. A phylogenomic study of birds reveals their evolutionary history. Science 320, 1763–1768 (2008)
Ericson, P. G. P. et al. Diversification of Neoaves: integration of molecular sequence data and fossils. Biology Letters 2, 543–547 (2006)
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012)
Prum, R. O. et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526, 569–573 (2015)
Thomas, G. H. & Freckleton, R. P. MOTMOT: models of trait macroevolution on trees. Methods Ecol. Evol. 3, 145–151 (2012)
Pennell, M. W. et al. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218 (2014)
Rabosky, D. L. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9, e89543 (2014)
Acknowledgements
We thank M. Adams, H. van Grouw and R. Prys-Jones from the Bird Group at the NHM, Tring and H. McGhie at the Manchester Museum for providing access to and expertise in the collections; S. Meiri of Tel Aviv University for providing a sample of study skins; S. Stone of MechInovation Ltd for providing training and advice on 3D scanning; M. Groves, J. McLaughlin and M. Pidd of HRI Digital for the construction of http://www.markmybird.org; A. Beckerman for advice on analysing P matrices; E. Rayfield, A. Pigot, A. Mooers and A. White for providing valuable comments on pre-submission drafts of the manuscript. Finally, we are indebted to the volunteer citizen scientists at http://www.markmybird.org for helping to build the database of bird bill shape and contribute to our understanding of avian evolution. This work was funded by the European Research Council (grant number 615709 Project ‘ToLERates’) and by a Royal Society University Research Fellowship to G.H.T. (UF120016).
Author information
Authors and Affiliations
Contributions
C.R.C., J.A.B. and G.H.T. conceived the study, designed analytical protocols, analysed the data and wrote the manuscript. All authors collected and processed data and provided input to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Rabosky and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Positions of landmarks and semi-landmarks.
The image shows a 3D scan of a shoebill (Balaeniceps rex) bill marked up with four fixed landmarks (numbered red points) and three semi-landmark curves along the dorsal profile (from points 1 to 2) and tomial edges (left from point 1 to 3 and right from point 1 to 4). Each curve consists of 25 semi-landmarks (black points).
Extended Data Figure 2 Morphospace density through time.
a–h, Plots show the filling of avian bill morphospace through time (n = 2,028 species) for PCs 1 (a), 2 (b), 3 (c), 4 (d), 5 (e), 6 (f), 7 (g) and 8 (h). Densities were calculated in 1-million-year time slices on the basis of univariate rate heterogeneous models of trait evolution using a stage 2 Hackett MCC tree from http://www.birdtree.org. The scale runs from low density (blue) to high density (red), indicating the extent of niche packing through time in different regions of bill morphospace. For each axis, the frequency distribution of PC scores among species is also shown (grey bars).
Extended Data Figure 3 Comparison of multivariate rates of bill-shape evolution and disparity through time for alternative datasets.
The plot shows estimates of the mean relative multivariate rate of bill-shape evolution for four alternative versions of the avian phylogeny and also when using phylogenetic principal components (pPCs) (see Methods). Shown below are plots comparing estimates of disparity and rates through time derived from each dataset. For stage 2 trees n = 2,028 species and for stage 1 trees n = 1,627 species.
Extended Data Figure 4 Multivariate rates of bill-shape evolution for a composite tree based on the Prum et al. backbone.
a, The avian phylogeny coloured according to estimates of the mean relative multivariate rate of bill-shape evolution. Grey triangles show the stem branch of clades with support for whole clade shifts in evolutionary rate. Coloured circles show rate shifts on individual internal branches (colour indicates the rate estimate). The relative size of triangles and circles indicates the posterior probability (PP) of a rate shift. Filled and open triangles distinguish between shifts on the focal node (filled) and shifts that occur either at the focal node or on one of the two immediate daughter nodes (open). b, Accumulation of multivariate disparity through time in 1 million year time slices (thick black line: observed data; thin black line: after LOESS smoothing; blue lines: constant rate null model; red lines: variable rate null model). c, Comparison of slopes (estimated in 5 million year windows) of the LOESS-smoothed observed data and null models. Differences in slope above and below zero indicate dominance of morphospace expansion versus morphospace packing, respectively. Shading indicates 95% confidence intervals. d, Mean relative rates of evolution with 95% confidence intervals (grey) through time.
Extended Data Figure 5 Phylogenetic mapping of univariate rates of bill-shape evolution.
The plots shows the avian phylogeny of all taxa included in the study (n = 2,028 species) with branches coloured on a common scale across panels according to estimates of the univariate rate of bill-shape evolution. a–h, PC1 (a), PC2 (b), PC3 (c), PC4 (d), PC5 (e), PC6 (f), PC7 (g) and PC8 (h).
Extended Data Figure 6 Morphospaces of avian higher taxa.
Pairwise scatter plots of PCs 1 and 2, 3 and 4, 5 and 6, and 7 and 8 showing focal higher taxa (non-passerines, purple; passerines, green) against total avian morphospace (grey). Values in parentheses show the number of species sampled.
Extended Data Figure 7 Morphological subspaces of the P of avian higher taxa.
The figure shows representations of P for avian higher taxa with ≥ 20 species sampled. First column: distribution of species values on each of the first eight raw PCs showing variation in morphospace centroid for each higher taxon. Second column: two-dimensional subspace for each taxon with non-passerine (purple) and passerine (green) subspaces. The x and y axes follow the global leading (Pmax) and secondary eigenvectors. Third column: percentage of total variance explained and individual PC loadings onto each taxon specific Pmax. Inset: 3D subspace for all non-passerines (purple) and passerines (green). Values in parentheses show the number of species sampled.
Supplementary information
Supplementary Data
This file details the mapping of Jetz et al. clades to the Prum et al. backbone phylogeny. The table shows the nodes used to attach patch clades from the Jetz et al. stage 2 Hackett tree to the Prum et al. backbone phylogeny. (XLSX 42 kb)
Supplementary Data
This archive contains data files and an R script to combine the backbone (approximately family level) phylogeny of Prum et al. with the species level resolution of the Jetz et al. avian phylogeny. (ZIP 794 kb)
Supplementary Data
This archive contains all alternative genus level phylogenies used in the analyses. (ZIP 253 kb)
Supplementary Data
This file contains the source data for Extended Data Table 1. (CSV 334 kb)
Rights and permissions
About this article
Cite this article
Cooney, C., Bright, J., Capp, E. et al. Mega-evolutionary dynamics of the adaptive radiation of birds. Nature 542, 344–347 (2017). https://doi.org/10.1038/nature21074
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21074
This article is cited by
-
Problem-solving skills are predicted by technical innovations in the wild and brain size in passerines
Nature Ecology & Evolution (2024)
-
Low morphological disparity and decelerated rate of limb size evolution close to the origin of birds
Nature Ecology & Evolution (2023)
-
Bird clades with less complex appendicular skeletons tend to have higher species richness
Nature Communications (2023)
-
Body size and life history shape the historical biogeography of tetrapods
Nature Ecology & Evolution (2023)
-
Allometry reveals trade-offs between Bergmann’s and Allen’s rules, and different avian adaptive strategies for thermoregulation
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.