Abstract
We are just beginning to understand how translational control affects tumour initiation and malignancy. Here we use an epidermis-specific, in vivo ribosome profiling strategy to investigate the translational landscape during the transition from normal homeostasis to malignancy. Using a mouse model of inducible SOX2, which is broadly expressed in oncogenic RAS-associated cancers, we show that despite widespread reductions in translation and protein synthesis, certain oncogenic mRNAs are spared. During tumour initiation, the translational apparatus is redirected towards unconventional upstream initiation sites, enhancing the translational efficiency of oncogenic mRNAs. An in vivo RNA interference screen of translational regulators revealed that depletion of conventional eIF2 complexes has adverse effects on normal but not oncogenic growth. Conversely, the alternative initiation factor eIF2A is essential for cancer progression, during which it mediates initiation at these upstream sites, differentially skewing translation and protein expression. Our findings unveil a role for the translation of 5′ untranslated regions in cancer, and expose new targets for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011)
Zhang, B. et al. Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 (2014)
Mamane, Y., Petroulakis, E., LeBacquer, O. & Sonenberg, N. mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 (2006)
Pelletier, J., Graff, J., Ruggero, D. & Sonenberg, N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 75, 250–263 (2015)
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009)
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012)
Huang, P. Y. & Balmain, A. Modeling cutaneous squamous carcinoma development in the mouse. Cold Spring Harb. Perspect. Med. 4, a013623 (2014)
Okubo, T., Pevny, L. H. & Hogan, B. L. M. Sox2 is required for development of taste bud sensory cells. Genes Dev. 20, 2654–2659 (2006)
Que, J. et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531 (2007)
Boumahdi, S. et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014)
Rudin, C. M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012)
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013)
Beronja, S. et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501, 185–190 (2013)
Beronja, S., Livshits, G., Williams, S. & Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16, 821–827 (2010)
Liu, K. et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell 12, 304–315 (2013)
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009)
Brar, G. A. et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2012)
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011)
Tyner, A. L. & Fuchs, E. Evidence for posttranscriptional regulation of the keratins expressed during hyperproliferation and malignant transformation in human epidermis. J. Cell Biol. 103, 1945–1955 (1986)
Signer, R. A. J., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014)
Blanco, S. et al. Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340 (2016)
Calviello, L. et al.. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170 (2016)
Slavoff, S. A. et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 (2013)
Falini, B. et al. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica 92, 519–532 (2007)
Yang, H. et al. ETS family transcriptional regulators drive chromatin dynamics and malignancy in squamous cell carcinomas. eLife 4, e10870 (2015)
Morris, D. R. & Geballe, A. P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20, 8635–8642 (2000)
Luo, B. et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl Acad. Sci. USA 105, 20380–20385 (2008)
Hinnebusch, A. G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 (2014)
Harding, H. P. et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000)
Skabkin, M. A. et al. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 24, 1787–1801 (2010)
Dmitriev, S. E. et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 285, 26779–26787 (2010)
Zoll, W. L., Horton, L. E., Komar, A. A., Hensold, J. O. & Merrick, W. C. Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem. 277, 37079–37087 (2002)
Terenin, I. M., Dmitriev, S. E., Andreev, D. E. & Shatsky, I. N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15, 836–841 (2008)
Starck, S. R. et al. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336, 1719–1723 (2012)
Starck, S. R. et al. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351, aad3867 (2016)
Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015)
Liu, B. & Qian, S.-B. Translational reprogramming in cellular stress response. WIREs RNA 5, 301–315 (2014)
Koromilas, A. E. Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim. Biophys. Acta 1849, 871–880 (2015)
Komar, A. A. et al. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J. Biol. Chem. 280, 15601–15611 (2005)
Reineke, L. C., Cao, Y., Baus, D., Hossain, N. M. & Merrick, W. C. Insights into the role of yeast eIF2A in IRES-mediated translation. PLoS One 6, e24492 (2011)
Holcik, M. Could the eIF2α-independent translation be the achilles heel of cancer? Front. Oncol. 5, 264 (2015)
Liang, H. et al.. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 19, 836–848 (2014)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Gerashchenko, M. V. & Gladyshev, V. N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 42, e134 (2014)
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protocols 7, 1534–1550 (2012)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Dunn, J. G. & Weissman, J. S. Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics 17, 958 (2016)
Beronja, S. & Fuchs, E. RNAi-mediated gene function analysis in skin. Methods Mol. Biol. 961, 351–361 (2013)
Platt, R. J. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014)
Faul, F., Erdfelder, E., Buchner, A. & Lang, A.-G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160 (2009)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Blanpain, C., Lowry, W. E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004)
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014)
Ishihama, Y., Rappsilber, J. & Mann, M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 (2006)
Kleifeld, O. et al. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat. Protocols 6, 1578–1611 (2011)
Hofacker, I. L. RNA secondary structure analysis using the Vienna RNA package. Curr. Protoc. Bioinformatics Chapter 12, Unit12.2 (2009)
Acknowledgements
We thank J. Que for the R26-Sox2-IRES-eGFP mice, D. Xu and the Cornell Genomics Facility for sequencing support, the Rockefeller Proteomics Facility for protein/peptide analyses, members of the Fuchs’ laboratory for discussions, and L. Polak and M. Sribour for their support with tumorigenesis studies. We thank E. Heller for bioinformatics support, L. Calviello for support with RiboTaper, and F. Garcia-Quiroz and M. Jovanovic for critical reading of the manuscript. The Rockefeller University Proteomics Resource Center acknowledges funding from the Leona M. and Harry B. Helmsley Charitable Trust and Sohn Conferences Foundation for mass spectrometer instrumentation. The results published here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). A.S. was supported by the Human Frontier Science Program Organization (HFSP, LT000639-2013) and is currently supported by the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7 under REA grant agreement no. 629861. S.N. is a Damon Runyon Fellow (DRG-2183-14). B.H. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. E.F. and J.S.W. are Investigators of the Howard Hughes Medical institute. This work was supported by grants to E.F. from the National Institutes of Health (R37-AR27883) and NYSTEM CO29559.
Author information
Authors and Affiliations
Contributions
A.S. and E.F. conceived the project, designed the experiments and wrote the manuscript. A.S. and B.H. performed the experiments, and collected and analysed data. J.G.D., E.H.R., J.S.W. and N.C.G. contributed to ribosome profiling data analysis. D.S. contributed to control shRNA library generation and established HrasG12V; Tgfbr2-null cell lines. S.N. contributed to OPP experiments. J.L. carried out in utero lentiviral injections. H.M. and B.D.D. performed proteomics experiments and analysed proteomics data. E.F. supervised the project. All authors discussed the results and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Consequence of SOX2 expression in the epidermis and correlation between in vivo ribosome profiling experiments.
a, Papilloma formation induced by SOX2 overexpression in the skin. SOX2 expression in tamoxifen-inducible K14-creER; R26-Sox2-IRES-eGFPfl/fl mice results in hyperplasia and papilloma formation. Representative H&E sections are shown. Animals develop severe skin lesions in the ventral epidermis 6–8 weeks after tamoxifen injection and require euthanasia. b, Experimental strategy to perform in vivo epidermis-specific ribosome profiling. In vivo epidermis-specific ribosome profiling strategy results in highly reproducible quantifications. Plots show RPKM correlations between the 3 independent replicate experiments in wild-type P4 epidermis. Quantified in this study were mRNAs with >128 reads. c, N-terminal extension of the translated Swi5 mRNA in wild-type and SOX2 P4 epidermis in vivo. Tracks show ribosome profiling reads along the Swi5 mRNA for replicate samples of each genotype. The final track shows harringtonine-treated ribosome profiling reads of wild-type keratinocytes in vitro. Harringtonine blocks ribosomes at the translational start site and allows translation start site mapping18. Red arrow indicates direction of translation. Green bar marks the annotated CDS. Blue bar denotes the actual translated coding sequence based upon ribosomal profiling.
Extended Data Figure 2 Global translational efficiency is decreased upon SOX2 expression in keratinocytes in vitro.
a, Ribosome profiling data correlates with proteomics data. At a stage when proliferation and morphology were similar, freshly isolated, basally enriched keratinocytes from wild-type and SOX2-expressing P4 skins were subjected to in vivo ribosome profiling and to a label-free proteomics strategy. Plotted are proteomics fold changes compared to ribosome profiling fold changes. Comparisons are made in SOX2 versus wild-type samples for significantly changed proteins (false discovery rate (FDR) < 0.05). b, Translational efficiency is markedly reduced in SOX2-expressing premalignant keratinocytes in vitro. Keratinocytes were isolated from R26-Sox2-IRES-eGFPfl/+ (WT) or K14-cre; R26-Sox2-IRES-eGFPfl/+ (SOX2+) P0 animals and cultured in vitro. Quantified were genes with more than 256 reads in RNA-seq data. Histogram shows distribution of differential translational efficiency (TE = RPKMribosome profiling/RPKMRNA-seq). Data are shown from 4 independent ribosome profiling experiments and 2 independent RNA-seq experiments. c, Wild-type and SOX2 keratinocytes have similar proliferation rates in vivo. Basal epidermal EdU incorporation in P0, P2 and P4 mice (n = 442/496, 385/449 and 903/841 cells from duplicate wild-type/SOX2 animals) 1 h after injection. Data are mean ± s.d. d, Phospho-4E-BP1 immunohistochemistry in the epidermis shows no difference in levels upon SOX2 induction. e, NSUN2 transcript, translation, and protein levels in SOX2 versus WT P4 epidermis.
Extended Data Figure 3 Translationally controlled genes show a shift towards cancer-related pathways.
Pathway analyses reveal a shift towards cancer-related pathways in mRNAs resistant to the SOX2-mediated global decrease in translational efficiency and in mRNAs with preferential uORF translation in premalignant SOX2+ P4 epidermis. Ingenuity pathway analysis (IPA) was used to analyse genes differentially regulated at four levels of control: transcriptional, translational, translational efficiency, and uORF/ORF ratio level. Included were the top 10% most upregulated and top 10% most downregulated genes at the levels of the transcriptome, translatome, and uORF usage. The translational efficiency list was restricted to the top and bottom-most 500 genes, corresponding to 6.6% of all genes. Total number of genes quantified: 4,725 transcriptome, 4,725 translatome, 7,605 translational efficiency, 1,830 uORFs.
Extended Data Figure 4 Translation from 5′ UTRs.
a, 5′ UTR translation of Myc in the P4 epidermis in vivo. As described previously, Myc mRNA contains several translated uORFs18. Right panels show higher magnification of the uORF start codons. b, 5′ UTR translation of Eif4g2 (encoding eIF4γ2) in the P4 epidermis in vivo. Harringtonine track shows main translation initiation site in the 5′ UTR. Right panels show higher magnification of the uORF start codons. c, 5′ UTR translation of Btg1 in the P4 epidermis in vivo. Harringtonine track shows main translation initiation site in the 5′ UTR. Right panels show higher magnification of the uORF start codons. d, Relative uORF translation in SOX2 versus wild-type keratinocytes using Harringtonine-treated samples. For each gene, the ratio of ribosome footprints in 5′ UTR of Harringtonine samples versus CHX-treated ORFs (for normalization) was calculated. Histogram shows distribution of log2 fold changes in relative 5′ UTR translation.
Extended Data Figure 5 Proteomic detection of 13 peptides produced from 5′ UTRs.
a, The RiboTaper analysis pipeline22 was used to annotate upstream ORFs computationally in an in vitro SOX2 keratinocyte sample. RiboTaper exploits the triplet periodicity of ribosomal footprints to predict bona fide translated regions. Boxplot shows the distribution of length of 215 uORFs which start with an AUG codon with a median length of 54 nucleotides. As a comparison, the right panel shows boxplot with the distribution of length of main ORFs predicted by RiboTaper. b, Schematic overview of the terminal amine isotopic labelling of substrates (TAILS) strategy to specifically enrich for N-terminal fragments. c, Overview of the pre- and post-TAILS peptide counts and peptide-spectrum matches. d, MS/MS spectra for identified uORF peptides. Representative MS/MS spectra for identified uORF peptides with monoisotopic (m/z), parent mass error (parts per million), charge state, and Mascot ion score. Matched y-ion fragments are shown in blue, b-ions in red, and unfragmented parents in green. Peptide N termini were identified as naturally N-terminally acetylated or unmodified. Owing to the protein-level primary amine blocking step in the TAILS workflow, naturally unmodified N termini and all lysines carry a dimethyl chemical modification. e, Ribosome profiling tracks showing translated uORF in P4 epidermis of the hepatoma-derived growth factor Hdgf and of the glycolipid transfer Gltp gene. Encoded peptides identified by high-resolution/high-mass-accuracy mass spectrometry using proteomics and peptidomics are shown. Red amino acids correspond to the identified peptides, yellow nucleotides mark potential initiation sites.
Extended Data Figure 6 SOX2-targeted 5′ UTRs are highly structured.
a, Genes with increased SOX2-regulated 5′ UTR translation also have longer 5′ UTRs and are more structured. The 10% of genes with the most increased 5′ UTR translation in SOX2 cells were evaluated relative to the 10% of genes with the largest decrease in 5′ UTR translation (error bars indicate range, n = 183 each, two-sided Wilcoxon test). b, Cumulative distribution plot showing length to structure comparison, an assessment for each gene’s 5′ UTR structure relative to its length. Analysis showing that SOX2-upregulated 5′ UTRs tend to have more favourable free energy at each length, suggesting that SOX2-regulated 5′ UTR secondary structures are more stable even when normalized for length (error bars indicate range, n = 183 each, two-sided Wilcoxon test). c, mRNAs showing preferential uORF translation in premalignant SOX2-expressing epidermis significantly overlap with mRNAs that are most resistant to the reduction in translational efficiency in premalignant SOX2 versus wild-type epidermis (hypergeometric test, P < 0.001). All mRNAs with a relative increase in the uORF/ORF ratio in SOX2 versus wild type were compared to the top 10% of mRNAs most resistant to reduction in translational efficiency in P4 SOX2 versus wild-type epidermis. Pathway analysis for the overlapping 122 genes (ingenuity pathway analysis) revealed epithelial–mesenchymal transition (EMT), stem-cell pluripotency, and axonal guidance as the top most enriched pathways. d, HrasG12V; Tgfbr2 knockout SCC tumour growth is dependent on SOX2 signalling. As shown in Fig. 4h, the HrasG12V; Tgfbr2 knockout is sufficient to upregulate SOX2 levels. Graph showing SCC tumour growth post-injection of 105 cells. Data are mean ± s.e.m. (n = 8 for each genotype). e, Overlap between uORF translation that occurs preferentially in premalignant SOX2-expressing P4 epidermis in vivo and uORF translation of malignant SOX2-expressing, HRAS-regulated SCC in vitro. Included were all mRNAs with twofold difference SOX2 versus wild-type P4 epidermis in vivo and twofold difference HrasG12V; Tgfbr2 knockout SCC versus wild-type keratinocytes in vitro. Hypergeometric test, P < 0.001.
Extended Data Figure 7 An shRNA screen reveals regulatory nodes in premalignancy.
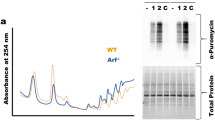
a, The 138 ribosomal genes and initiation factors targeted in our shRNA screen. b, Overview of the shRNA screen in wild-type versus premalignant SOX2-expressing epidermis from P0 mice. The RIGER algorithm27 was used with the following methods and metrics to convert hairpins to genes and to rank top hits. From left to the right: weighted sum, signal to noise (median); second best rank, signal to noise (median); weighted sum, signal to noise; weighted sum, fold change. c, Western blot shows knockdown efficiency of shRNAs targeting the top hit in our screen, Eif2s1. Note that the knockdown efficiency correlates well with the degree of shRNA depletion in the screen. d, Intra-amniotic injection of lentivirus library of Eif2a and 35 control shRNAs. This sub-library was injected into E9.5 embryos and representation of shRNAs was quantified in wild-type and SOX2 P0 skin. Top, knockdown efficiency; bottom, relative representation of Eif2a shRNAs (normalized against the control shRNA library) in wild-type versus SOX2 epidermis. e, DNA sequence of Eif2a knockout clonal cell lines used in our study. PAM region is highlighted in red, CRISPR target region in blue. f, g, Proliferation rates and overall protein synthesis rates are unchanged in Eif2a knockout cells under serum-rich conditions in vitro. SCC control and Eif2a knockout cells were quantified for EdU and OPP incorporation 1 h after administration. Data are mean ± s.d. of 4 independent experiments. h, The translational landscape is unchanged in Eif2a knockout cells under serum-rich conditions in vitro. SCC control and Eif2a knockout cells subjected to ribosome profiling and reads within the main ORFs were quantified and tested for differential expression using DESeq2. As opposed to 5′ UTR translation, no significant differences (adjusted P < 0.1 DESeq2) in ORF translation were found between SCC control and SCC Eif2a knockout cells (n = 2 SCC control, n = 2 SCC Eif2a knockout). Right panels show two representative H&E sections of squamous cell carcinomas formed 25 days after subcutaneous injection of SCC cells. i, eIF2A-dependent changes of heavy-labelled peptides of CTNNB1 and CD44 under stress in pulsed SILAC. SCC control and SCC Eif2a knockout cells were grown in light-labelled medium and switched to heavy-labelled medium supplemented with 5 μM arsenite. Graphs show the relative difference between SCC control and SCC Eif2a knockout cells during the pulsed SILAC time course.
Extended Data Figure 8 mRNAs containing eIF2A-targeted uORFs are preferentially translated during tumorigenesis.
a–c, Genes that contain eIF2A-targeted uORFs maintain increased translation and translational efficiency of their downstream ORFs during early stages of tumorigenesis. HrasG12V; Tgfbr2 knockout SCCs were subcutaneously injected into nude mice. Day 5 or day 10 tumours were analysed by ribosome profiling and RNA-seq. Changes in transcription, translation and translational efficiency were assessed comparing in vivo against SCC in vitro data and represented as fold changes in Kernel density plots. Either only the significantly changed genes (DESeq2) or all genes were assessed. eIF2A-targeted versus non-eIF2A-targeted uORF genes refer to changes in uORF usage in SCC control versus SCC Eif2a knockout in Fig. 6a (n = 2 SCC in vitro, n = 2 SCC day 5, n = 2 SCC day 10, two-sample Kolmogorov–Smirnov test). d, Mutations in the eIF2A-regulated uORF of Ctnnb1, a key oncogene for SCC progression, diminishes its tumorigenic potential. Tracks show ribosome profiling reads in the 5′ UTR of Ctnnb1 and in the uORF GUG start codon. HrasG12V; Tgfbr2 knockout SCC keratinocytes were infected with either non-targeting control sgRNA or Ctnnb1 uORF sgRNAs. Clonal lines were sequenced and an uORF mutant clonal line was established. Data are mean ± s.e.m. following subcutaneous injection of 105 cells (n = 8 control, n = 14 clone 2B).
Extended Data Figure 9 Human EIF2A is frequently amplified in human cancer.
a, Summary of cross-cancer alterations for EIF2A in human cancers12. 29% of patients with lung carcinoma and 15% of patients with head and neck SCCs show an amplification of the EIF2A locus. b, Summary of the clinical information accompanying TCGA patients with head and neck SCC.
Extended Data Figure 10 Overview of 5′ UTR translation and proteomics analyses.
a, Summary of different analyses of 5′ UTR translation in this study. b, Significantly changed proteins in SOX2 versus wild-type P4 epidermis in vivo (FDR < 0.05).
Supplementary information
Supplementary Figure
This file contains the raw data for Figures 4h and 5a,d. (PDF 94 kb)
Source data
Rights and permissions
About this article
Cite this article
Sendoel, A., Dunn, J., Rodriguez, E. et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499 (2017). https://doi.org/10.1038/nature21036
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21036
This article is cited by
-
Beyond genetics: driving cancer with the tumour microenvironment behind the wheel
Nature Reviews Cancer (2024)
-
Nucleoporin downregulation modulates progenitor differentiation independent of nuclear pore numbers
Communications Biology (2023)
-
Genome-wide identification of Arabidopsis non-AUG-initiated upstream ORFs with evolutionarily conserved regulatory sequences that control protein expression levels
Plant Molecular Biology (2023)
-
Principles, challenges, and advances in ribosome profiling: from bulk to low-input and single-cell analysis
Advanced Biotechnology (2023)
-
Non-AUG translation initiation in mammals
Genome Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.