Abstract
Cell fate perturbations underlie many human diseases, including breast cancer1,2. Unfortunately, the mechanisms by which breast cell fate are regulated are largely unknown. The mammary gland epithelium consists of differentiated luminal epithelial and basal myoepithelial cells, as well as undifferentiated stem cells and more restricted progenitors3,4. Breast cancer originates from this epithelium, but the molecular mechanisms that underlie breast epithelial hierarchy remain ill-defined. Here, we use a high-content confocal image-based short hairpin RNA screen to identify tumour suppressors that regulate breast cell fate in primary human breast epithelial cells. We show that ablation of the large tumour suppressor kinases (LATS) 1 and 2 (refs 5, 6), which are part of the Hippo pathway, promotes the luminal phenotype and increases the number of bipotent and luminal progenitors, the proposed cells-of-origin of most human breast cancers. Mechanistically, we have identified a direct interaction between Hippo and oestrogen receptor-α (ERα) signalling. In the presence of LATS, ERα was targeted for ubiquitination and Ddb1–cullin4-associated-factor 1 (DCAF1)-dependent proteasomal degradation. Absence of LATS stabilized ERα and the Hippo effectors YAP and TAZ (hereafter YAP/TAZ), which together control breast cell fate through intrinsic and paracrine mechanisms. Our findings reveal a non-canonical (that is, YAP/TAZ-independent) effect of LATS in the regulation of human breast cell fate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Derynck, R. & Akhurst, R. J. Differentiation plasticity regulated by TGF-β family proteins in development and disease. Nat. Cell Biol. 9, 1000–1004 (2007)
Tong, Q. & Hotamisligil, G. S. Developmental biology: cell fate in the mammary gland. Nature 445, 724–726 (2007)
Howard, B. A. & Gusterson, B. A. Human breast development. J. Mammary Gland Biol. Neoplasia 5, 119–137 (2000)
Petersen, O. W. & Polyak, K. Stem cells in the human breast. Cold Spring Harb. Perspect. Biol. 2, a003160 (2010)
Halder, G. & Johnson, R. L. Hippo signaling: growth control and beyond. Development 138, 9–22 (2011)
Zhao, B., Li, L., Lei, Q. & Guan, K. L. The Hippo–YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 (2010)
Shaw, F. L. et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J. Mammary Gland Biol. Neoplasia 17, 111–117 (2012)
Villadsen, R. et al. Evidence for a stem cell hierarchy in the adult human breast. J. Cell Biol. 177, 87–101 (2007)
Lim, E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913 (2009)
Santagata, S. et al. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J. Clin. Invest. 124, 859–870 (2014)
Regan, J. L. et al. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 31, 869–883 (2012)
Lim, E. et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 12, R21 (2010). 10.1186/bcr2560
Charafe-Jauffret, E. et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 25, 2273–2284 (2006)
Doane, A. S. et al. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25, 3994–4008 (2006)
Chen, Q. et al. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 28, 432–437 (2014)
Skibinski, A. et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Reports 6, 1059–1072 (2014)
Bhat, K. P. et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 25, 2594–2609 (2011)
Chen, D. et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo–YAP pathway and a prognostic marker. Nat. Med. 18, 1511–1517 (2012)
Cordenonsi, M. et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011)
Harvey, K. F., Zhang, X. & Thomas, D. M. The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013)
Balwierz, P. J. et al. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 24, 869–884 (2014)
Siegel, P. M. & Muller, W. J. Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene 29, 2753–2759 (2010)
Shehata, M. et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14, R134 (2012)
Matthews, L. et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37, D619–D622 (2009)
Zhang, J., Smolen, G. A. & Haber, D. A. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 68, 2789–2794 (2008)
Zhang, J. et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 11, 1444–1450 (2009)
Varelas, X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141, 1614–1626 (2014)
Linnemann, J. R. et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development 142, 3239–3251 (2015)
Li, W. et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4DCAF1-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 26, 48–60 (2014)
Gyorffy, B., Lánczky, A. & Szállási, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 19, 197–208 (2012)
St John, M. A. et al. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21, 182–186 (1999)
Couse, J. F. & Korach, K. S. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev. 20, 358–417 (1999)
Duss, S. et al. Mesenchymal precursor cells maintain the differentiation and proliferation potentials of breast epithelial cells. Breast Cancer Res. 16, R60 (2014)
Stampfer, M., Hallowes, R. C. & Hackett, A. J. Growth of normal human mammary cells in culture. In Vitro 16, 415–425 (1980)
Stingl, J., Emerman, J. T. & Eaves, C. J. Enzymatic dissociation and culture of normal human mammary tissue to detect progenitor activity. Methods Mol. Biol. 290, 249–263 (2005)
König, R. et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods 4, 847–849 (2007)
Duss, S. et al. An oestrogen-dependent model of breast cancer created by transformation of normal human mammary epithelial cells. Breast Cancer Res. 9, R38 (2007)
Britschgi, A. et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 22, 796–811 (2012)
Sleeman, K. E., Kendrick, H., Ashworth, A., Isacke, C. M. & Smalley, M. J. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 8, R7 (2006)
Cicalese, A. et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138, 1083–1095 (2009)
Meier-Abt, F. et al. Parity induces differentiation and reduces Wnt/Notch signaling ratio and proliferation potential of basal stem/progenitor cells isolated from mouse mammary epithelium. Breast Cancer Res. 15, R36 (2013)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Soule, H. D. et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 (1990)
Bentires-Alj, M. et al. A role for the scaffolding adapter GAB2 in breast cancer. Nat. Med. 12, 114–121 (2006)
Ignatiadis, M. & Sotiriou, C. Luminal breast cancer: from biology to treatment. Nat. Rev. Clin. Oncol. 10, 494–506 (2013)
Koren, S. et al. PIK3CAH1047R induces multipotency and multi-lineage mammary tumours. Nature 525, 114–118 (2015)
Brauchle, M. et al. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One 8, e60549 (2013)
Pomp, V. et al. Differential expression of epithelial–mesenchymal transition and stem cell markers in intrinsic subtypes of breast cancer. Breast Cancer Res. Treat. 154, 45–55 (2015)
Varga, Z., Tubbs, R. R. & Moch, H. Concomitant detection of HER2 protein and gene alterations by immunohistochemistry (IHC) and silver enhanced in situ hybridization (SISH) identifies HER2 positive breast cancer with and without gene amplification. PLoS One 9, e105961 (2014)
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012)
Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11, 367 (2010)
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) Method. Methods 25, 402–408 (2001)
Acknowledgements
We thank members of the Bentires-Alj laboratory for advice and discussions. P. Ringenbach, S. Thiebault and V. Lindner provided mammary tissue. We were supported by FMI and GNF facilities: S. Thiry for microarray hybridizations, H. Kohler for FACS analysis, S. Bichet for histology, L. Gelmant, R. Thierry, K. Volkmann and S. Bourke for imaging, A. Romero for lentiviral preparations, B. Tu for automation of the screen, and K. Drumm for animal studies. We thank T. Radimerski, A. Bauer, T. Schmelzle and M. Frederiksen for discussions and reagents. Research in the laboratory of M.B.-A. is supported by the Novartis Research Foundation, the European Research Council, the Swiss National Science Foundation, the Krebsliga Beider Basel, the Swiss Cancer League, the Department of Surgery of the University Hospital of Basel and the Swiss Initiative for Systems Biology (SystemsX.ch).
Author information
Authors and Affiliations
Contributions
A.B., S.D. and S.Ki. conceived the study, designed and performed experiments, analysed the data, interpreted the results and wrote the manuscript. J.P.C. performed PHBECs and MECs FACS and imaging, lentiviral infection, cell culture and limiting dilution transplantations, analysed the data and interpreted the results. H.B. performed immunohistochemistry and immunofluorescence experiments, analysed the data and interpreted the results. D.D.S. and S.Ko. performed MECs FACS, cell culture, immunofluorescence and limiting dilution transplantations. K.D.M., D.K. and Z.V. performed and analysed experiments on archived human breast cancer samples. H.V., A.V. and C.L. performed immunoprecipitation and mass spectrometry experiments and analysed the data. T.R. and M.B.S. performed microarray data analysis. C.H.S. interpreted 3D collagen assay experiments. G.M.C.B. imaged and analysed the 3D imaging data and the resulting screen data. L.J.M. and A.P.O. contributed to the shRNA screen experiment and analysis. V.A.R. and M.B.-A. conceived the study, designed the experiments and interpreted the results. M.B.-A. wrote and V.A.R. revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
J.P.C., D.D.S., K.D.M., D.K., Z.V., T.R., M.B.S., S.Ko., C.H.S. and M.B.-A. declare no competing financial interests. A.B., S.D. and H.B. are employees of Roche Pharma AG. S.Ki. is an employee of Amgen. H.V., A.V., L.M., A.P.O. and G.M.C.B. are employees of Novartis Pharma AG. C.L. is an employee of Actelion. V.A.R. is a full-time employee of Pfizer.
Additional information
Reviewer Information Nature thanks C. Watson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
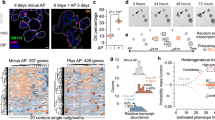
Extended Data Figure 1 Inhibition of LATS increases sphere formation and the fractions of progenitor and luminal cells.
a, Outline of the screen to assess effects of the inhibition of tumour suppressor genes on PHBEC cell fate. b, Representative immunofluorescence images of shNT-, shLATS1- and shLATS2-treated PHBEC spheres stained for KRT14 (red) and KRT19 (green). Scale bar, 50 μm. c, Top, bar graphs showing fractions of KRT14+KRT19+ (left axis) and curve showing the ratio of KRT19+ to KRT14+ (right axis) cells upon inhibition of tumour suppressor genes; the top 40 individual shRNAs are shown. shRNAs targeting components of the Hippo pathway are indicated by red arrows. Bottom, bar graphs representing fractions of KRT14+, KRT19+ and KRT14+KRT19+ cells in spheres (n = 5 single shRNAs, 4 experimental replicates). d, Inhibition of Lats increases sphere formation in mouse primary mammary epithelial cells (MECs). Immunoblots and bar graph showing sphere formation of shNT and shLats cells (n = 25 pooled mice, 6 experimental replicates). e, Representative immunofluorescence images of shNT- and shLATS1 + 2-treated PHBEC colonies stained for KRT14 (red) and KRT19 (green). Scale bars, 150 μm. f, Knockdown of LATS enhances sphere formation in MCF10A cells. Immunoblots (left) and bar graph (right) showing sphere formation by shNT- and shLATS-treated cells cultured in M5 medium (n = 6 experimental replicates). g, The fraction of KRT14+ basal cells decreased, whereas the fractions of KRT18+ luminal and KRT14+KRT18+ progenitor cells increased upon removal of LATS. Representative immunofluorescence images of shNT- and shLATS-treated MCF10A colonies stained for KRT14 and KRT18 (left) and corresponding quantification (right) (n = 6 experimental replicates). Scale bars, 150 μm. h, LATS knockdown increases KRT19 and decreases KRT14 mRNA expression in PHBECs and MCF10A cells. qPCR analysis of shNT- and shLATS-treated cells (n = 3 biological replicates, and 2 and 6 experimental replicates each for PHBECs and MCF10A cells, respectively). i, Re-expression of wild-type LATS1 rescues the effects of the knockdown in MCF10A cells. Immunoblots (left), representative immunofluorescence pictures (middle) and corresponding quantification of cells treated with shNT or shLATS1C (targeting the 3′UTR) that were transfected with either control or LATS1 wild-type cDNA-expressing vectors (n = 6 experimental replicates). Scale bars, 200 μm. Data are mean ± s.d. (c, d, f–i); *P < 0.05; **P < 0.01.
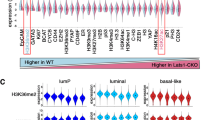
Extended Data Figure 2 Inhibition of LATS promotes a luminal phenotype.
a, Heat map derived from hierarchical clustering of the 182 genes that were differentially expressed between shNT-treated PHBECs (n = 5) and shLATS-treated PHBECs (n = 6). Cut-off: adjusted P < 0.01, fold change > 2.0, expression values > 4. Arrows indicate canonical luminal genes as well as LATS1. b, Principal component analysis (PCA) of microarrays performed on shNT- and shLATS-treated PHBECs. shNT-treated samples cluster separately from both shLATS1B- and shLATS1 + 2-treated samples. c, Venn diagram analysis of differentially regulated genes at a low stringency cut-off of P < 0.05, 2.0-fold change, expression values > 4.0. Both LATS shRNAs show a high degree of overlap when compared to shNT-treated samples and no genes were significantly differentially regulated between cells treated with the two shRNAs. d, shLATS-treated PHBEC expression profiles were significantly enriched in genes that are downregulated in normal basal breast cells. GSEA with shLATS-specific genes and a gene set of normal basal/stem breast cells. shNT-treated PHBECs (n = 5), shLATS-treated PHBECs (n = 6). NES = 2.5; FDR < 0.0001; P < 0.0001. e, shLATS-treated PHBECs express low levels of basal genes. Heatmaps with average normalized expression values of a subset of genes associated with normal basal breast cells. f, Removal of LATS imposes a luminal breast cancer signature on PHBECs. Left, GSEA with shLATS-specific genes derived from PHBECs and gene sets of ERα-positive (left graph) and luminal breast cancer samples (right graph). shNT-treated PHBECs (n = 5), shLATS-treated PHBECs (n = 6). NES = 2.29 (left) and 1.98 (right); FDR <0.0001; P < 0.0001. Right, clustered heatmap showing correlation coefficients between human breast cancer profiles and shNT- or shLATS-treated PHBECs. g, Graphs of ISMARA transcription factor activity analysis of shLATS- (n = 6) versus shNT-treated (n = 5) PHBECs showing high activity upon removal of LATS for CEBPβ (Z = 1.92; P < 0.0001) and STAT5A,B (Z = 2.184, P < 0.01). h, qPCR analysis of shNT- and shLATS-treated PHBECs. mRNA levels of ERα and c-KIT as well as several canonical ERα-target genes are higher in shLATS-treated PHBECs (n = 6 experimental and 2 technical replicates), i, FACS analysis of PHBECs with LATS knockdown. Markers of luminal progenitors (c-KIT, ALDH activity) and luminal epithelial (EPCAM) cells increased and expression of the basal marker CD10 was reduced upon removal of LATS (n = 6 experimental replicates). Data are mean ± s.d. (h, i); *P < 0.05, **P < 0.01; NS, not significant.
Extended Data Figure 3 YAP/TAZ are stabilized in shLATS cells but do not impose their canonical signature.
a, YAP/TAZ signalling increases upon removal of LATS. Immunoblots (left) and qPCR analysis (right) of shNT- and shLATS-treated PHBECs. Data are mean ± s.d. (n = 6 experimental replicates), *P < 0.05. b, GSEA analysis of the shLATS profiles with datasets in which YAP was overexpressed (left and middle) or with a YAP/TAZ gene set derived from http://www.reactome.org/ (right). Left, NES = 0.84; FDR = 0.726; P = 0.726. Middle, NES = 0.91; FDR = 0.58; P = 0.58. Right, NES = 0.1.17; FDR = 0.251; P = 0.251. There was no enrichment of the shLATS-treated PHBEC profile with the published YAP- or YAP/TAZ-driven gene set.
Extended Data Figure 4 LATS-depleted MCF10A cells recapitulate the phenotype of LATS-depleted PHBECs.
a, MCF10A cells lacking LATS are enriched for luminal gene signatures but not for YAP/TAZ-driven gene signatures. GSEA with shLATS-treated MCF10A cell-specific genes and gene sets from PHBECs lacking LATS (left), luminal progenitor (middle) and mature luminal breast cells (right). Results from enrichment analyses with YAP/TAZ-driven gene sets are shown in the table. b, Markers of mature luminal and luminal progenitor cells increased upon removal of LATS, and canonical YAP/TAZ signalling was activated. Immunoblots of lysates (left) and bar graphs showing qPCR measurements (middle), and FACS analysis (right) of shNT- and shLATS-treated MCF10A cells cultured in standard growth medium (Std) or M5 medium for 4 days. c, Left, representative microscopic brightfield pictures of shNT- and shLATS-treated MCF10A cells cultured in M5 medium showing a switch to a cobblestone-like cell morphology in shLATS-treated cells. Scale bars, 150 μm. Right, qPCR analysis of shNT- and shLATS-treated MCF10A cells. Luminal markers are upregulated and mesenchymal markers are downregulated in shLATS-treated MCF10A cells. Data are mean ± s.d.; n = 6 experimental and 2 technical replicates; *P < 0.05; **P < 0.01; NS, not significant.
Extended Data Figure 5 YAP/TAZ and ERα signalling promote sphere formation and breast luminal phenotype in LATS-depleted cells via intrinsic and paracrine mechanisms.
a, Cells with enforced expression of both YAP/TAZ and ERα phenocopy shLATS-treated cells. Immunoblots (left) of shNT-treated, shNT-treated with enforced YAP/TAZ and/or ERα expression and shLATS-treated MCF10A cells. PCA plots (middle) and heatmap (right) derived from hierarchical clustering of genes that were differentially expressed between shNT-treated, shNT-treated YAP/TAZ expressing, shNT-treated ERα expressing, shNT-treated ERα and YAP/TAZ expressing, shLATS1B-treated and shLATS1 + 2-treated MCF10A cells (n = 3 experimental replicates). Cut-off: adjusted P < 0.01; fold change > 2.0; expression values > 4. b, GSEA showing that cells with exogenous YAP/TAZ expression are enriched for YAP/TAZ signatures. c, Immunoblots representing siRNA-mediated depletion of YAP/TAZ in shNT- and shLATS-treated MCF10A cells (left). Inhibition of YAP/TAZ only partially blocks the induction of luminal markers (qPCR, right), but reverses enhanced sphere formation upon LATS knockdown (middle) (n = 8 experimental replicates). d, Immunoblots representing siRNA-mediated depletion of ERα in MCF10A shNT- and shLATS-treated cells (left). Inhibition of ERα blocks the induction of luminal markers (qPCR, right), but only slightly reduces the enhanced sphere formation seen upon LATS knockdown (middle) (n = 8 experimental replicates). e, Inhibition of ERα by 1 μM 4-hydroxytamoxifen (4-OHT) in PHBECs blocks the induction of luminal markers (qPCR, left, n = 3 experimental and 2 technical replicates), but only slightly reduces the enhanced sphere formation seen upon LATS knockdown (right, n = 6 experimental replicates). f, Immunoblots of shNT- and shLATS-treated MCF10A cells in which YAP, TAZ and ERα were depleted by siRNA. g, YAP/TAZ and ERα interact in the upregulation of AREG in cells lacking LATS. Left, ELISA measurements in shNT- and shLATS-treated PHBECs and MCF10A cells (n = 8 experimental replicates). Right, qPCR results for shNT- and shLATS-treated MCF10A cells in which YAP/TAZ and/or ERα were depleted by siRNA (n = 6 experimental and 2 technical replicates). h, Left, sphere formation by shNT- and shLATS-treated MCF10A cells upon addition of increasing amounts of an AREG-blocking antibody (AB) or an IgG control. n = 6 experimental replicates; IC50 values are shNT = 8.2 μg; shLATS1 = 4.4 μg (P < 0.05); shLATS_2 = 2.6 μg (P < 0.05); shLATS_3 = 2.7 μg (P < 0.05). Right, sphere formation by MCF10A cells supplemented with conditioned medium derived from either shNT- or shLATS-treated MCF10A 3D cultures. Addition of anti-AREG blocking antibody (2.5 μg ml−1) reversed the increase in sphere-forming capacity conferred by conditioned medium from shLATS-treated cultures (n = 6 experimental replicates). i, Graphical summary. Data are mean (h (left)) or mean ± s.d. (c–e, g, h (right)); *P < 0.05; NS, not significant.
Extended Data Figure 6 Removal of LATS increases proliferation and favours survival of ERα+ cells.
a, Knockdown of LATS induces markers of proliferation (immunoblots) and increases cell numbers of PHBECs (cell counts) (n = 5 experimental and 2 technical replicates). b, Knockdown of LATS increases colony-forming capacity (colony counts) of PHBECs (n = 8 experimental replicates). c, Knockdown of LATS increases the expression of markers of proliferation. Immunoblots of MCF10A cells with or without LATS cultured in M5 medium. d, Knockdown of LATS increases cell number. Cell counts of shNT- and shLATS-treated MCF10A cells cultured in M5 medium (n = 8 experimental replicates). e, shLATS-treated cells in suspension retain high levels of YAP/TAZ and are partially anoikis-resistant. Immunoblots of shNT- and shLATS-treated MCF10A cells cultured in suspension for the indicated time points (left). FACS analysis of annexin V+ and propidium iodide+ MCF10A cells after 36 h of suspension culture (right) (n = 8 experimental replicates). f, Removal of LATS results in a survival advantage for ERα+ cells. Scheme of the setup of the competition experiment (left). Immunoblots of lysates from shNT- and shLATS-treated MCF10A cells after lentiviral infection with GFP and ERα or the empty vector control at the beginning of the experiment. MOIs for ERα were adjusted between shNT- and shLATS-treated cell lines to assure equal expression of ERα (middle). FACS analysis of GFP+ cells after 5 days of suspension culture (right) (n = 6 experimental replicates). g, Inhibition of YAP/TAZ partially reverses the competitive advantage of ERα+ shLATS-treated cells. The experimental setup was as in f, but in addition YAP/TAZ were inhibited by siRNA. Immunoblots of lysates of shNT- and shLATS-treated MCF10A cells at the beginning of the experiment, infected with GFP and ERα, and transfected with siRNAs against YAP/TAZ, and a non-targeting control (left). FACS analysis of GFP+ cells after 5 days of suspension culture (right) (n = 6 experimental replicates). Data are mean (a) or mean ± s.d. (b, e–g); *P < 0.05, **P < 0.01.
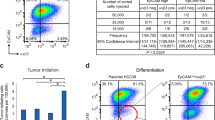
Extended Data Figure 7 Removal of LATS increases the luminal progenitor and mature luminal cell phenotype.
a, LATS is mostly nuclear in basal cells and cytoplasmic in luminal cells. Representative pictures of LATS1 staining (immunohistochemistry) in normal human breast (left; scale bar, 100 μm), of PHBECs stained (immunofluorescence) with KRT19, KRT14 and LATS1 (middle; scale bars, 150 μm), and of a human breast tumour TMA (right; scale bars, 50 μm). Quantification of LATS nuclear and cytoplasmic staining by immunofluorescence in PHBECs (middle, n = 8). Box plots representing ERα-index of tumours characterized by cytoplasmic or nuclear LATS immunohistochemistry staining (right; n = 471; P < 0.0001). b, qPCR analysis of sorted PHBEC subpopulations (fold change over stroma and mesenchymal cells (MSC)). Luminal markers are shown in blue, basal markers are shown in red (n = 2 biological replicates with 3 experimental replicates each). c, The fractions of mature luminal and luminal progenitor cells increase upon removal of LATS. Representative scatter plots and bar graphs of cell-surface/intracellular FACS analysis of shNT- and shLATS-treated PHBECs stained with antibodies against c-KIT and ERα (n = 2 biological replicates with 3 experimental replicates each). d, Representative immunofluorescence pictures of colonies formed by breast cell subpopulations. Scale bars, 150 μm. e, YAP/TAZ targets are highly expressed in basal cells. qPCR analysis of sorted basal and c-KIT+ luminal progenitor cells (n = 2 biological replicates with 3 experimental replicates each). f, Sorted basal primary mouse MECs acquire a luminal phenotype upon removal of Lats, and removal of Lats enhances the luminal phenotype in sorted luminal MECs. Representative immunofluorescence pictures and quantification of the colony phenotype of sorted basal (top) and luminal (bottom) MECs with and without Lats1 + 2 stained for KRT14 and KRT8/18. n = 20 pooled mice; basal shNT, n = 46 colonies; basal shLats, n = 23 colonies; luminal shNT, n = 30 colonies; luminal shLats, n = 20 colonies. Scale bars, 200 μm. g, Depletion of Lats in sorted primary MECs increases luminal mature and progenitor markers and decreases basal and mammary stem-cell-related markers (qPCR; n = 20 pooled mice; 6 experimental replicates). h, Cells lacking LATS exhibit a luminal progenitor-like phenotype when grown in 3D collagen gels. Left, representative pictures of TDLU- and sphere-like structures formed by PHBECs on floating collagen. Right, representative immunofluorescence confocal pictures of 3D structures stained for basal (KRT14) and luminal (KRT18, E-cadherin) markers. Scale bars, 200 μm. i, Depletion of Lats in primary mouse MECs increases luminal mature and progenitor markers and decreases basal and mammary stem-cell-related markers (qPCR, n = 25 pooled mice, 6 experimental replicates). j, Graphical summary. Depletion of LATS expands luminal and bipotent progenitors and enhances differentiation along the luminal lineage at the expense of basal progenitors and basal cells. This results in a population of cells with high sphere-forming capacity, low mammary repopulating activity, and a luminal phenotype. Data are mean ± s.d.(a–c, e, g, i); *P < 0.05; **P < 0.01.
Extended Data Figure 8 Removal of LATS stabilizes ERα protein.
a, Knockdown of LATS upregulates ERα in luminal breast cancer cells. Immunoblots of lysates of shNT- and shLATS-treated T47D cells treated for 6 h with 10 nM oestrogen (E2) or ethanol (EtOH). b, Knockdown of LATS upregulates ERα-target genes in luminal breast cancer cells. qPCR analysis of shNT- and shLATS-treated T47D cells treated as in a. c, Re-expression of the kinase-dead LATS1 mutant almost fully rescues ERα-mediated effects of the knockdown in MCF10A cells, but not YAP/TAZ-mediated effects. Immunoblots (top left), quantification of sphere formation (top right) and qPCR analysis (bottom) of MCF10A cells with or without LATS cDNAs expressing LATS1 wild-type (WT) or mutant (mut) sequences. d, ERα protein is stabilized in cells lacking LATS. Immunoblots of lysates from shNT- and shLATS-treated T47D cells that were treated with 10 μM of the protein-translation inhibitor cycloheximide (CHX) and 10 nM oestrogen for 6 h. Quantification of ERα levels, normalized to loading controls ERK2 and β-actin and expressed relative to 0 h. e, LATS1 triggers proteasome-dependent downregulation of ERα protein. Immunoblots of lysates from BT474 and MCF7 cells transfected with wild-type LATS1 cDNA and treated with 20 μM of the proteasome inhibitor MG132 for 6 h. f, g, Ubiquitination of ERα is reduced in shLATS-treated cells and is dependent on DCAF1. Immunoblots of MCF7 shNT and shLATS cell lysates (f) and MCF7 cells transfected with LATS1 wild-type cDNA and siDCAF1 (g) subjected to ERα immunoprecipitation and immunoblotting for ubiquitin and ERα. IP, immunoprecipitation; WCL, whole cell lysate. b–d, Data are mean ± s.d.; n = 6 experimental replicates; *P < 0.05; NS, not significant.
Extended Data Figure 9 LATS ablation reduces the efficacy of fulvestrant treatment.
a, LATS is downregulated in the majority of human breast cancer samples and cell lines. Immunoblots of PHBECs and primary human breast cancer samples (top), MCF10A cells and basal (bottom left) and luminal (bottom right) breast cancer cell lines. b, shLATS reduces sensitivity to fulvestrant by preventing ERα degradation. Colony areas of T47D (top) and of MCF7 (bottom) shNT- and shLATS-treated cell lines in response to fulvestrant and tamoxifen treatment. (n = 9 experimental replicates). c, shLATS-treated tumours are partially resistant to fulvestrant. Tumour growth curves of shNT- and shLATS-treated T47D cell-bearing animals treated with 2.5 mg fulvestrant per 25 g, as indicated n = 5 biological replicates. d, Low LATS expression is associated with poor outcome in patients treated with anti-oestrogens but not in the tamoxifen-only treated subgroup. Kaplan–Meier analysis of relapse-free survival of n = 643 (left), n = 346 (middle), n = 489 (right) patients with breast cancer. Data are mean ± s.d. (b, c); *P < 0.05.
Extended Data Figure 10 Graphical summary.
Effects of LATS in luminal and basal cells. In luminal cells, LATS is highly expressed in the cytoplasm, where it triggers YAP/TAZ degradation. These cells express high levels of ERα and its target genes. In basal cells, LATS is highly expressed in the nucleus, allowing YAP/TAZ accumulation in the cytoplasm and translocation to the nucleus, where they activate their targets. Upon depletion of LATS, YAP/TAZ are upregulated in luminal cells with the net results that both ERα and YAP/TAZ target genes are being transcribed. In basal cells, removal of LATS results in stabilization of ERα and the transcription of ERα target genes in addition to the already highly expressed YAP/TAZ target genes. Overall, these changes result in a cell population characterized by increases in the levels of luminal genes, in the number of luminal and bipotent progenitors, in sphere formation, and proliferation.
Supplementary information
Supplementary Figure
This file contains the raw data for Figures 1b, 2c, 4b,d,e. (PDF 345 kb)
Supplementary Table 1: shRNA tumor suppressor library used for the 3D screen
TRC numbers, oligo sequences, clone names, reference sequences, and gene names of the shRNA constructs used for the 3D cell fate screen in PHBECs. (XLSX 32 kb)
Supplementary Table 2: Raw data of the 3D screen
Excel file containing results of K14 and K19 immunofluorescent staining as well as morphological parameters (number and size of spheres) of all shRNA conditions. Marker 1 = K14, Marker 2 = K19. (XLSX 442 kb)
Supplementary Table 3: Results of redundant shRNA analysis (RSA) of the 3D screen
Excel file containing results of RSA analysis of the number of spheres as well as the number of double-positive cells. (XLSX 738 kb)
Supplementary Table 4: Differentially regulated genes in PHBECs upon removal of LATS
Excel file of the significantly differentially regulated genes in the contrast shLATS versus shNT at a cut-off of fold-change (FC) >2.0, expression value >4.0, adjusted P <0.01. (XLSX 39 kb)
Supplementary Table 5: Differentially regulated genes in MCF10A cells upon removal of LATS and/or expression of YAP, TAZ and ERα
Excel file of the significantly differentially regulated genes at a cut-off of fold-change (FC) >2.0, expression value >4.0, adjusted P <0.01. (XLSX 116 kb)
Supplementary Table 6: ERα interactome in luminal breast cancer cells with and without LATS1
Excel file of ratios of ERα interacting proteins in the presence or absence of LATS1. (XLSX 95 kb)
Rights and permissions
About this article
Cite this article
Britschgi, A., Duss, S., Kim, S. et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα. Nature 541, 541–545 (2017). https://doi.org/10.1038/nature20829
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20829
This article is cited by
-
LATS1 controls CTCF chromatin occupancy and hormonal response of 3D-grown breast cancer cells
The EMBO Journal (2024)
-
Palbociclib sensitizes ER-positive breast cancer cells to fulvestrant by promoting the ubiquitin-mediated degradation of ER-α via SNHG17/Hippo-YAP axis
Breast Cancer Research and Treatment (2024)
-
DNMT3a-dermatopontin axis suppresses breast cancer malignancy via inactivating YAP
Cell Death & Disease (2023)
-
The oncogenic roles and clinical implications of YAP/TAZ in breast cancer
British Journal of Cancer (2023)
-
Deep learning-based system for automatic prediction of triple-negative breast cancer from ultrasound images
Medical & Biological Engineering & Computing (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.