Abstract
Metastasis is the leading cause of death for cancer patients. This multi-stage process requires tumour cells to survive in the circulation, extravasate at distant sites, then proliferate; it involves contributions from both the tumour cell and tumour microenvironment (‘host’, which includes stromal cells and the immune system1). Studies suggest the early steps of the metastatic process are relatively efficient, with the post-extravasation regulation of tumour growth (‘colonization’) being critical in determining metastatic outcome2. Here we show the results of screening 810 mutant mouse lines using an in vivo assay to identify microenvironmental regulators of metastatic colonization. We identify 23 genes that, when disrupted in mouse, modify the ability of tumour cells to establish metastatic foci, with 19 of these genes not previously demonstrated to play a role in host control of metastasis. The largest reduction in pulmonary metastasis was observed in sphingosine-1-phosphate (S1P) transporter spinster homologue 2 (Spns2)-deficient mice. We demonstrate a novel outcome of S1P-mediated regulation of lymphocyte trafficking, whereby deletion of Spns2, either globally or in a lymphatic endothelial-specific manner, creates a circulating lymphopenia and a higher percentage of effector T cells and natural killer (NK) cells present in the lung. This allows for potent tumour cell killing, and an overall decreased metastatic burden.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nature Med. 19, 1423–1437 (2013)
Chambers, A. F. et al. Critical steps in hematogenous metastasis: an overview. Surg. Oncol. Clin. N. Am. 10, 243–255 (2001)
Blank, C. et al. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 64, 1140–1145 (2004)
van Elsas, A., Hurwitz, A. A. & Allison, J. P. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med. 190, 355–366 (1999)
White, J. K. et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154, 452–464 (2013)
Ogasawara, K. et al. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature 391, 700–703 (1998)
Honda, K. et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777 (2005)
Purtha, W. E., Swiecki, M., Colonna, M., Diamond, M. S. & Bhattacharya, D. Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc. Natl Acad. Sci. USA 109, E898–E904 (2012)
Sasaki, T. et al. Function of PI3Kγ in thymocyte development, T cell activation, and neutrophil migration. Science 287, 1040–1046 (2000)
Tassi, I. et al. p110γ and p110δ phosphoinositide 3-kinase signaling pathways synergize to control development and functions of murine NK cells. Immunity 27, 214–227 (2007)
Kitamura, D., Roes, J., Kühn, R. & Rajewsky, K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature 350, 423–426 (1991)
Sun, X. et al. CD39/ENTPD1 expression by CD4+Foxp3+ regulatory T cells promotes hepatic metastatic tumor growth in mice. Gastroenterology 139, 1030–1040 (2010)
Guerrero, J. A. et al. Gray platelet syndrome: proinflammatory megakaryocytes and α-granule loss cause myelofibrosis and confer metastasis resistance in mice. Blood 124, 3624–3635 (2014)
Okada, F. et al. The role of nicotinamide adenine dinucleotide phosphate oxidase-derived reactive oxygen species in the acquisition of metastatic ability of tumor cells. Am. J. Pathol. 169, 294–302 (2006)
Kang, B. H. et al. Targeted inhibition of mitochondrial Hsp90 suppresses localised and metastatic prostate cancer growth in a genetic mouse model of disease. Br. J. Cancer 104, 629–634 (2011)
Takabe, K. & Spiegel, S. Export of sphingosine-1-phosphate and cancer progression. J. Lipid Res. 55, 1839–1846 (2014)
Fukuhara, S. et al. The sphingosine-1-phosphate transporter Spns2 expressed on endothelial cells regulates lymphocyte trafficking in mice. J. Clin. Invest. 122, 1416–1426 (2012)
Mendoza, A. et al. The transporter Spns2 is required for secretion of lymph but not plasma sphingosine-1-phosphate. Cell Reports 2, 1104–1110 (2012)
Nagahashi, M. et al. Spns2, a transporter of phosphorylated sphingoid bases, regulates their blood and lymph levels, and the lymphatic network. FASEB J. 27, 1001–1011 (2013)
Wilkerson, B. A. & Argraves, K. M. The role of sphingosine-1-phosphate in endothelial barrier function. Biochim. Biophys. Acta 1841, 1403–1412 (2014)
Chiang, E. Y., Henson, M. & Stroynowski, I. Correction of defects responsible for impaired Qa-2 class Ib MHC expression on melanoma cells protects mice from tumor growth. J. Immunol. 170, 4515–4523 (2003)
Schwab, S. R. et al. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science 309, 1735–1739 (2005)
Nijnik, A. et al. The role of sphingosine-1-phosphate transporter Spns2 in immune system function. J. Immunol. 189, 102–111 (2012)
Garris, C. S., Blaho, V. A., Hla, T. & Han, M. H. Sphingosine-1-phosphate receptor 1 signalling in T cells: trafficking and beyond. Immunology 142, 347–353 (2014)
Walzer, T. et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nature Immunol. 8, 1337–1344 (2007)
Cuff, S., Dolton, G., Matthews, R. J. & Gallimore, A. Antigen specificity determines the pro- or antitumoral nature of CD8+ T cells. J. Immunol. 184, 607–614 (2010)
Ponnusamy, S. et al. Communication between host organism and cancer cells is transduced by systemic sphingosine kinase 1/sphingosine 1-phosphate signalling to regulate tumour metastasis. EMBO Mol. Med. 4, 761–775 (2012)
Visentin, B. et al. Validation of an anti-sphingosine-1-phosphate antibody as a potential therapeutic in reducing growth, invasion, and angiogenesis in multiple tumor lineages. Cancer Cell 9, 225–238 (2006)
Liu, G., Yang, K., Burns, S., Shrestha, S. & Chi, H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nature Immunol. 11, 1047–1056 (2010)
Liu, Y. et al. The sphingosine-1-phosphate receptor agonist FTY720 and its phosphorylated form affect the function of CD4+CD25+ T cells in vitro. Int. J. Mol. Med. 30, 211–219 (2012)
Pham, T. H. et al. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J. Exp. Med. 207, 17–27 (2010)
Shinkai, Y. et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992)
Kranz, A. et al. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis 48, 512–520 (2010)
Karp, N. A. et al. Impact of temporal variation on design and analysis of mouse knockout phenotyping studies. PLoS ONE 9, e111239 (2014)
Kilkenny, C., Browne, W. J., Cuthill, I. C., Emerson, M. & Altman, D. G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 (2010)
Borsig, L., Wong, R., Hynes, R. O., Varki, N. M. & Varki, A. Synergistic effects of L- and P-selectin in facilitating tumor metastasis can involve non-mucin ligands and implicate leukocytes as enhancers of metastasis. Proc. Natl Acad. Sci. USA 99, 2193–2198 (2002)
Johnstone, C. N. et al. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis. Model. Mech . 8, 237–251 (2015)
Bald, T. et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature 507, 109–113 (2014)
Lindsay, C. R. et al. P-Rex1 is required for efficient melanoblast migration and melanoma metastasis. Nature Commun . 2, 555 (2011)
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature Methods 10, 1096–1098 (2013)
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols 9, 171–181 (2014)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015)
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Hait, N. C. et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325, 1254–1257 (2009)
Curran, P. J. & Hussong, A. M. Integrative data analysis: the simultaneous analysis of multiple data sets. Psychol. Methods 14, 81–100 (2009)
Hochberg, Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75, 800–802 (1988)
Acknowledgements
This work was supported by grants from Cancer Research UK (D.J.A. and O.J.S.), the Wellcome Trust (WT098051), Combat Cancer (D.J.A.), the European Research Council (311301 COLONCAN to O.J.S. and A.D.C.), National Institutes of Health U54HG004028 (N.A.K.), and Department of Defense BCRP Program Award W81XWH-14-1-0086 (S.S.). T.T. was funded by project A27N in the SFB854, and T.B. was funded in part by an EMBO Long-Term Fellowship (ALTF 945-2015) and the European Commission (Marie Curie Action LTFCOFUND2013, GA-2013-609409). We thank J. Allegood for sphingolipid analyses and acknowledge the VCU Lipidomics Core, which is supported in part by funding from the National Institutes of Health–National Cancer Institute (NIH–NCI) Cancer Center Support Grant P30CA016059, V. Iyer (Wellcome Trust Sanger Institute) for bioinformatics analysis, and members of the Wellcome Trust Sanger Institute Research Support Facility for their care of the mice.
Author information
Authors and Affiliations
Consortia
Contributions
L.v.d.W. devised and implemented the pulmonary metastasis screen, performing all the primary screen, confirmation and characterization studies. M.J.A. analysed the histopathological sections. A.D.C. and O.J.S. performed and analysed the intrasplenic B16-F10 assays. T.B. and T.T. performed and analysed the spontaneous metastasis assay. H.W.-J. and N.G. managed mouse breeding and were responsible for issuing phenotyping cohorts. M.D.C.V.-H., T.V., I.C.M. and K.W. performed the RNA-seq analysis. D.G. and E.R. genotyped the mice and performed gene expression analysis. S.C., A.G., E.T. and E.L.C. performed additional phenotypic characterization. The Sanger Mouse Genetics Project generated and phenotyped the mice as part of a primary phenotyping pipeline. S.S. oversaw the lipidomic analysis and provided input to the project and the manuscript. A.O.S. devised, performed and analysed the immunophenotyping assays. L.v.d.W., A.O.S. and D.J.A. led the project. L.v.d.W., A.O.S. and D.J.A. wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information: Nature thanks C. Ghajar and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Lists of participants and their affiliations appear in the Supplementary Information
Extended data figures and tables
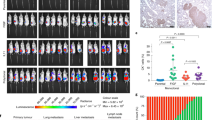
Extended Data Figure 1 Molecular function of 810 mutant mouse lines screened and phenotypic characterization members of the interferon regulatory factor (Irf) family.
a, Molecular function Gene Ontology annotation of the 810 mutant mouse lines screened as detailed in Methods. b, Experimental metastasis assay using B16-F10 cells in Irf1tm1a/tm1a, Irf5tm1e/tm1e, Irf7tm1a/tm1a and concurrent control female mice. Shown are representative data from two (Irf5), four (Irf1) or six (Irf7) independent experiments. Symbols represent individual mice with a horizontal bar at the mean. P values are from a Mann–Whitney test. c–f, Representative photographs showing B16-F10 metastatic colonies on the (c) lungs of +/+ and Irf1tm1a/tm1a mice and (d–f) the presence of extra-pulmonary metastases in Irf1tm1a/tm1a mice (tissues from three mice shown).
Extended Data Figure 2 Spontaneous pulmonary metastases and primary tumour growth in Spns2 mice.
a, Size measurements of spontaneous pulmonary HCmel12–mCherry melanoma cell metastases of male mice with representative fluorescent images (lines indicate the edge of the lungs); n = 10 per genotype, horizontal bars represent mean (of 50 individual metastases counted per genotype) (one-way ANOVA with blocking factor of experiment, cumulative results of two independent experiments shown). b, Survival curve of +/+ and tm1a/tm1a male mice (n = 10 per genotype) in a spontaneous metastasis assay using HCmel12–mCherry cells (log-rank test (Mantel–Cox), cumulative results of two independent experiments shown). c, Growth of subcutaneously administered B16-BL6 cells in +/+ (four male, five female) and tm1a/tm1a (five male, one female) mice. Symbols represent mean ± s.e.m. with a two-tailed unpaired t-test with Welch’s correction used to compare the area under the curve. d, Incidence of cancer in aged (>40 weeks) +/+ (n = 15; 4 males, 11 females) and tm1a/tm1a (n = 18; 5 males, 13 females) mice. Statistical analysis was performed using a Fisher’s exact test.
Extended Data Figure 3 Phenotyping of the serum and lungs of Spns2 mice.
Sphingoid base levels in the (a) serum (+/+, n = 5; tm1a/tm1a, n = 4) and (b) lungs (+/+, n = 6; tm1a/tm1a, n = 5) of male mice; data are mean ± s.e.m., multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%. Sph, sphingosine; DHSph, dihydrosphingosine; S1P, sphingosine-1-phosphate; DHS1P, dihydrosphingosine-1-phosphate. c, Micrograms of extravasated Evans blue dye in the lungs of +/+ and tm1a/tm1a male mice. d, Number of CFSE-labelled B16-F10 cells present in the lungs of female mice 90 min after administration. e, Levels of apoptosis in B16-F10–mCherry cells 12 h after administration to male mice. Shown are representative data from three independent experiments, with symbols representing individual mice. P values are indicated from two-tailed unpaired t-test with Welch’s correction (c–e).
Extended Data Figure 4 Phenotypic characterization of the haematopoietic system of Spns2 mice.
a–c, The numbers of erythrocytes and platelets, monocytes, granulocytes and lymphocyte subsets present in the blood of naive +/+ and tm1a/tm1a female mice (multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%; data shown are representative of three independent experiments). d, Analysis of lymphocyte subsets in the liver of naive +/+ and tm1a/tm1a female mice (multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%; data shown are representative of three independent experiments). e, f, T- and B-lymphocyte numbers in the blood of male naive (unstimulated) bone marrow chimaeras (unpaired two-tailed t-test with Welch’s correction; data shown are representative of two independent experiments). Symbols represent individual mice; horizontal bars represent mean.
Extended Data Figure 5 Characterization of the phenotype of lymphatic endothelial cell Spns2 deficient mice.
a, b, Sphingoid base levels in the (a) serum or (b) lung of control and Spns2tm1c/tm1c; Lyve1cre/+ male mice (data are mean ± s.e.m., control n = 11, Spns2tm1c/tm1c; Lyve1cre/+ n = 10, multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%). Sph, sphingosine; DH-Sph, dihydrosphingosine; S1P, sphingosine-1-phosphate; DH-S1P, dihydrosphingosine-1-phosphate. c, Lymphocyte subsets in the spleen, lymph node, lung and liver of +/+ and tm1a/tm1a male mice (symbols represent individual mice, horizontal bars represent mean, multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%; data shown are representative of three independent experiments). d, Experimental metastasis assay using MC-38 cells in control (n = 9) and Spns2tm1c/tm1c; Lyve1cre/+ (n = 5) in female mice. Data shown are mean ± s.e.m., Mann–Whitney test, representative of three independent experiments.
Extended Data Figure 6 T cell subsets in the lungs of Spns2 mice.
The proportion of T cell subsets present in the lungs of naive +/+ and tm1a/tm1a female mice (a, b, e) and control and Spns2tm1c/tm1c; Lyve1cre/+ male mice (c, d, f). Data are shown as percentage of parent CD4+ and CD8+ T cells (a, c, e, f) or percentage of CD45+ alive lung cells present (b, d). Symbols represent individual mice with horizontal bar at the mean. P values are indicated from two-tailed unpaired t-test adjusted by the Holm–Šídák method with α set to 5%. Data shown are representative of three independent experiments.
Extended Data Figure 7 T cell subsets in the liver of Spns2 mice.
The proportion of T cell subsets present in the liver of naive +/+ versus tm1a/tm1a female mice and control versus Spns2tm1c/tm1c; Lyve1cre/+ male mice. Data are shown as percentage of parent CD4+ and CD8+ T cells (a, c, e, f) or percentage of CD45+ alive liver cells present (b, d). Symbols represent individual mice; statistical analysis used multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%, with * indicating a P value not considered significant after correcting for multiple testing. Data shown are representative of three independent experiments.
Extended Data Figure 8 Phenotyping of Spns2 lungs.
a, b, Ex vivo re-stimulation (PMA/ionomycin) of pulmonary leukocytes from B16-F10-stimulated +/+ and tm1a/tm1a female mice (two-tailed unpaired t-test adjusted by the Holm–Šídák method with α set to 5%). c, Measurement of IFN-γ in lungs of MC-38-stimulated +/+ and tm1a/tm1a male mice (two-tailed unpaired t-test with Welch’s correction). d, e, The proportion of NK cell subsets present in the lungs of naive +/+ versus tm1a/tm1a female mice (d) and control versus Spns2tm1c/tm1c; Lyve1cre/+ male mice (e) (multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%). Symbols represent individual mice, horizontal bars represent mean; data shown are representative of three independent experiments.
Extended Data Figure 9 Studies in T- and B-cell-deficient mice.
a, Measurement of lymphocyte subsets in the blood of +/+ and Rag2−/− mice (multiple two-tailed unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%). b, Experimental metastasis assay using B16-F10 cells in +/+ and Rag2−/− female mice (Mann–Whitney test). Symbols represent individual mice, horizontal bars represent mean; data shown are representative of three independent experiments.
Extended Data Figure 10 Characterization of the leukocyte composition and phenotype in DOP-treated mice.
a–d, The number of leukocytes and T cell subsets present in the lungs of B16-F10-dosed glucose- or DOP-treated wild-type male mice presented as the percentages of viable CD45+ lung leukocytes (a, c) or parent CD4+ or CD8+ T cells (b, d) (multiple unpaired t-tests with P value adjusted by the Holm–Šídák method with α set to 5%). e, Experimental metastasis assay in B16-F10 dosed glucose- or DOP-treated wild-type female mice (Mann–Whitney test). Symbols represent individual mice, horizontal bars represent mean; data shown are representative of two independent experiments.
Supplementary information
Supplementary Information
This file contains a list of the Sanger Mouse Genetics Project participants and Supplementary Tables 1-4. (PDF 451 kb)
Source data
Rights and permissions
About this article
Cite this article
van der Weyden, L., Arends, M., Campbell, A. et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature 541, 233–236 (2017). https://doi.org/10.1038/nature20792
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20792
This article is cited by
-
Lipids as mediators of cancer progression and metastasis
Nature Cancer (2024)
-
Dissecting metastasis using preclinical models and methods
Nature Reviews Cancer (2023)
-
Targeting USP2 regulation of VPRBP-mediated degradation of p53 and PD-L1 for cancer therapy
Nature Communications (2023)
-
Signaling pathways in cancer metabolism: mechanisms and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
-
Iron Deficiency Increases Phosphorylation of SP1 to Upregulate SPNS2 Expression in Hepatocellular Carcinoma
Biological Trace Element Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.