Abstract
The functionality of stem cells declines during ageing, and this decline contributes to ageing-associated impairments in tissue regeneration and function1. Alterations in developmental pathways have been associated with declines in stem-cell function during ageing2,3,4,5,6, but the nature of this process remains poorly understood. Hox genes are key regulators of stem cells and tissue patterning during embryogenesis with an unknown role in ageing7,8. Here we show that the epigenetic stress response in muscle stem cells (also known as satellite cells) differs between aged and young mice. The alteration includes aberrant global and site-specific induction of active chromatin marks in activated satellite cells from aged mice, resulting in the specific induction of Hoxa9 but not other Hox genes. Hoxa9 in turn activates several developmental pathways and represents a decisive factor that separates satellite cell gene expression in aged mice from that in young mice. The activated pathways include most of the currently known inhibitors of satellite cell function in ageing muscle, including Wnt, TGFβ, JAK/STAT and senescence signalling2,3,4,6. Inhibition of aberrant chromatin activation or deletion of Hoxa9 improves satellite cell function and muscle regeneration in aged mice, whereas overexpression of Hoxa9 mimics ageing-associated defects in satellite cells from young mice, which can be rescued by the inhibition of Hoxa9-targeted developmental pathways. Together, these data delineate an altered epigenetic stress response in activated satellite cells from aged mice, which limits satellite cell function and muscle regeneration by Hoxa9-dependent activation of developmental pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rando, T. A. Stem cells, ageing and the quest for immortality. Nature 441, 1080–1086 (2006)
Brack, A. S. et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807–810 (2007)
Carlson, M. E. et al. Relative roles of TGF-β1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell 8, 676–689 (2009)
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014)
Conboy, I. M., Conboy, M. J., Smythe, G. M. & Rando, T. A. Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577 (2003)
Price, F. D. et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat. Med. 20, 1174–1181 (2014)
Krumlauf, R. Hox genes in vertebrate development. Cell 78, 191–201 (1994)
Lawrence, H. J., Sauvageau, G., Humphries, R. K. & Largman, C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells 14, 281–291 (1996)
Cosgrove, B. D. et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20, 255–264 (2014)
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999)
Lyons, K. M., Pelton, R. W. & Hogan, B. L. Organogenesis and pattern formation in the mouse: RNA distribution patterns suggest a role for bone morphogenetic protein-2A (BMP-2A). Development 109, 833–844 (1990)
Muñoz-Espín, D. et al. Programmed cell senescence during mammalian embryonic development. Cell 155, 1104–1118 (2013)
Sehgal, P. B., Levy, D. E. & Hirano, T. Signal Transducers And Activators Of Transcription (STATS): Activation And Biology (Kluwer Academic, 2003)
Sinha, M. et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344, 649–652 (2014)
Bernet, J. D. et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 20, 265–271 (2014)
Soshnikova, N. & Duboule, D. Epigenetic temporal control of mouse Hox genes in vivo . Science 324, 1320–1323 (2009)
Ayton, P. M. & Cleary, M. L. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9 . Genes Dev. 17, 2298–2307 (2003)
Yu, B. D., Hess, J. L., Horning, S. E., Brown, G. A. & Korsmeyer, S. J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995)
Grebien, F. et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 11, 571–578 (2015)
McKinnell, I. W. et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 10, 77–84 (2008)
Feller, C., Forné, I., Imhof, A. & Becker, P. B. Global and specific responses of the histone acetylome to systematic perturbation. Mol. Cell 57, 559–571 (2015)
Liu, L. et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Reports 4, 189–204 (2013)
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012)
Tierney, M. T. et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 20, 1182–1186 (2014)
Chakkalakal, J. V., Jones, K. M., Basson, M. A. & Brack, A. S. The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360 (2012)
Blagosklonny, M. V. Aging is not programmed: genetic pseudo-program is a shadow of developmental growth. Cell Cycle 12, 3736–3742 (2013)
Martin, N., Beach, D. & Gil, J. Ageing as developmental decay: insights from p16INK4a . Trends Mol. Med . 20, 667–674 (2014)
Lawrence, H. J. et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 89, 1922–1930 (1997)
Brown, P. J. & Müller, S. Open access chemical probes for epigenetic targets. Future Med. Chem. 7, 1901–1917 (2015)
Barsyte-Lovejoy, D. et al. Chemical biology approaches for characterization of epigenetic regulators. Methods Enzymol . 574, 79–103 (2016)
Theodoulou, N. H. et al. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 59, 1425–1439 (2016)
Picaud, S. et al. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 75, 5106–5119 (2015)
Picaud, S. et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 73, 3336–3346 (2013)
Martin, L. J. et al. Structure-based design of an in vivo active selective BRD9 inhibitor. J. Med. Chem. 59, 4462–4475 (2016)
Hay, D. A. et al. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains. J. Am. Chem. Soc. 136, 9308–9319 (2014)
Drouin, L. et al. Structure enabled design of BAZ2-ICR, a chemical probe targeting the bromodomains of BAZ2A and BAZ2B. J. Med. Chem. 58, 2553–2559 (2015)
Clark, P. G. et al. LP99: discovery and synthesis of the first selective BRD7/9 bromodomain inhibitor. Angew. Chem. 127, 6315–6319 (2015)
Chen, P. et al. Discovery and characterization of GSK2801, a selective chemical probe for the bromodomains BAZ2A and BAZ2B. J. Med. Chem. 59, 1410–1424 (2016)
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010)
Pasut, A., Jones, A. E. & Rudnicki, M. A. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. 73, 50074 (2013)
Bentzinger, C. F. et al. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell 12, 75–87 (2013)
Schambach, A. et al. Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp. Hematol. 34, 588–592 (2006)
Wang, J. et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148, 1001–1014 (2012)
Chambeyron, S. & Bickmore, W. A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 (2004)
Burgess, A. et al. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl Acad. Sci. USA 107, 12564–12569 (2010)
Opgen-Rhein, R. & Strimmer, K. Accurate ranking of differentially expressed genes by a distribution-free shrinkage approach. Stat. Appl. Genet. Mol. Biol. 6, http://dx.doi.org/10.2202/1544-6115.1252 (2007)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 12 (1995)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res . 32, D493–D496 (2004)
Shen, W. F. et al. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19, 3051–3061 (1999)
Huang, Y. et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 119, 388–398 (2012)
Acknowledgements
We thank Y. Morita and A. Illing for providing guidance regarding FACS analysis. We are thankful to the FLI Core Facilities Functional Genomics (T. Kroll, A. Ploubidou) and DNA Sequencing (M. Groth) for their services. We express our thanks to M. Burkhalter, T. Sperka and A. Illing for discussions and suggestions. We are grateful to B. Wollscheid and S. Goetze for providing support for proteomic measurements. We thank V. Sakk and M. Kettering for mouse husbandry as well as S. Eichwald, K. Tramm and A. Abou Seif for experimental assistance. We are grateful to M. Kessel, M. Kyba, G. Sauvageau and D. Wellik for sharing plasmids with Hox cDNAs. We thank the Structural Genomics Consortium and S. Ackloo for providing access to the epigenetic probe library. We further thank M. Cerletti for providing protocols on SC isolation and E. Perdiguero for advice on infection of SCs before transplantation. Work on this project in K.L.R.’s laboratory was supported by the DGF (RU-745/10, RU-745/12), the ERC (2012-AdG 323136), the state of Thuringia, and intramural funds from the Leibniz association. J.V.M. was supported by a grant from the DFG (MA-3975/2-1). C.F. acknowledges support by the DFG (FE-1544/1-1) and EMBO (long-term postdoctoral fellowship ALTF 55-2015). R.A. was supported by the ERC (AdvGr 670821 (Proteomics 4D)). The funding for the Hoxa9−/− mice to K.L.M. was provided by a grant of the NIH (HL096108). R.R. was supported by a grant from the NIH (R01GM106056). This work was further supported by grants to H.A.K. from the DFG (SFB 1074 project Z1), the BMBF (Gerontosys II, Forschungskern SyStaR, project ID 0315894A), and the European Community’s Seventh Framework Programme 390 (FP7/2007-2013, grant agreement 602783).
Author information
Authors and Affiliations
Contributions
S.S. designed and performed most experiments, analysed data, interpreted results and wrote the manuscript. F.B. designed and performed RNAi, ChIP and FISH experiments on isolated SCs, analysed data, interpreted results and wrote the manuscript. C.F. and R.A. designed and performed LC–MS experiments, analysed data, interpreted results and wrote the manuscript. A.H.B., U.K., H.H., C.S.V. and M.S. performed individual experiments and analysed data. A.L. performed microarray experiments. D.B.L. provided support and suggestions for ChIP experiments. K.L.M. provided Hoxa9−/− mice. J.M.K. and H.A.K. performed microarray and pathway analysis, analysed putative Hoxa9-binding sites and provided support for statistical analysis. B.X. and R.R. conducted analysis of putative Hoxa9-binding sites. F.N. analysed RNA-sequencing data and performed correlation analysis. J.V.M. and S.T. conceived the project, designed and performed individual experiments, interpreted results and wrote the manuscript. K.L.R. conceived the project, designed experiments, interpreted results and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks J. Gil and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
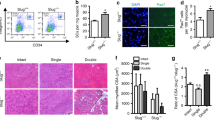
Extended Data Figure 1 SC activation.
a, Immunofluorescence staining for Pax7 and MyoD of freshly isolated SCs from injured (activated SCs) and uninjured muscles (quiescent SCs) from young adult mice. Nuclei were counterstained with DAPI (blue). b, c, Quantification of Pax7+ cells (b) and MyoD+ cells (c) in a. d, e, qPCR analysis of Spry1 (d) and Myod1 (e) expression in freshly isolated quiescent and in vivo activated SCs of young adult and aged mice. f, Immunofluorescence staining for Pax7 and MyoD on freshly isolated and 24-h cultured myofibre-associated SCs from aged mice. Nuclei were counterstained with DAPI (blue). g, Corrected total cell fluorescence (CTCF) for MyoD per SC as in f. Scale bars, 10 μm (a) and 20 μm (f). P values were calculated by two-sided Student’s t-test (b, c) or two-way ANOVA (d, e, g). n = 2 mice in b; n = 4 mice in c; n = 3 mice (young activated), n = 4 mice (all others) in d; n = 4 mice in e; n = 33/24 nuclei (young), n = 35/20 nuclei (aged) from 3 mice in g.
Extended Data Figure 2 Expression of Hox genes in SCs.
a, b, Nanostring analysis of mRNA expression of Hoxa genes and Hoxa9 paralogues (b9-c9-d9) in in vivo activated (a) and quiescent (b) freshly isolated SCs from young adult and aged mice. c, Relative fluorescence units (RFU) for Hoxa9 per SC in 4-day cultured SCs from young adult and aged mice. d, Corrected total cell fluorescence (CTFC) for Hoxa9 per activated SC on 24-h cultured myofibres as in Fig. 1d. P values were calculated by two-way ANOVA (a, b) or two-sided Mann–Whitney U-test (c, d). n = 3 mice in a, b; n = 3 mice (young), n = 5 mice (aged) in c; n = 34 nuclei (young), n = 32 nuclei (aged) from 4 mice in d.
Extended Data Figure 3 Functional decline in aged SCs.
a, SCs from young adult and aged mice were sorted as single cells. After 5 days, the frequency of myogenic colonies was assessed. b, Equal numbers of FACS-isolated SCs from young adult and aged mice were cultured for 4 days and Alamar Blue assay was performed. c, TUNEL staining of SCs isolated from young adult or aged mice after 4 days of culture. Nuclei were counterstained with DAPI (blue). d, Quantification of apoptosis based on TUNEL staining in c. e, BrdU staining of SCs isolated from young adult or aged mice after 4 days of culture. Nuclei were counterstained with DAPI (blue). f, Quantification of proliferation based on BrdU staining in e. g, Immunofluorescence staining for Pax7 and MyoD on myofibres isolated from young adult and aged mice after 72 h in culture. Nuclei were counterstained with DAPI (blue). h–j, Quantification of the number of SC-derived clusters with at least 3 adjacent cells (h), average number of all Pax7+ cells (i), or proportion of Pax7+/MyoD− cells (j) within clusters as in g. Scale bars, 20 μm (c, g) and 50 μm (e). P values were calculated by two-sided Student’s t-test. n = 8 mice (young), n = 10 mice (aged) in a; n = 7 mice (young), n = 5 mice (aged) in b; n = 3 mice in d; n = 4 mice in f; n = 4 mice (aged) in j, n = 5 mice (all others) in h–j.
Extended Data Figure 4 Deletion or knockdown of Hoxa9 improves SC function in myofibre cultures.
a, Immunofluorescence staining for Pax7 and MyoD on 72 h cultured myofibre-associated SCs from aged Hoxa9+/+ and Hoxa9−/− mice. b, c, Average number of all Pax7+ cells (b) or Pax7−/MyoD+ cells (c) within clusters from aged or young adult Hoxa9+/+ and Hoxa9−/− mice as shown in a. d, Immunofluorescence staining for Pax7 and MyoD on 72-h cultured myofibres isolated from aged mice transfected with Hoxa9 or scrambled (Scr) siRNAs. Nuclei were counterstained with DAPI (blue). e, qPCR analysis of Hoxa9 expression in SCs transfected with Hoxa9 siRNA or scrambled control. Two Hoxa9 siRNAs with different target sequences (Supplementary Table 1) were used. f–h, Analysis of 72-h cultured myofibre-associated SCs from d. Quantification of the number of SC-derived clusters with at least 3 adjacent cells (f), average number of all Pax7+ cells (g), or proportion of Pax7+/MyoD− cells (h) within clusters. Scale bars, 20 μm (a, d). Dashed lines outline myofibres. P values were calculated by two-sided Student’s t-test. n = 3 mice (aged), n = 4 mice (young) in b, c; n = 3 mice in e; n = 5 mice in f–h.
Extended Data Figure 5 Inhibition of Hoxa9 improves muscle regeneration in aged mice.
a, Quantification of Pax7+ cells per area in uninjured tibialis anterior muscles from young adult and aged Hoxa9+/+ and Hoxa9−/− mice. b, Representative immunofluorescence staining for Pax7 and laminin on tibialis anterior muscles from aged Hoxa9+/+ and Hoxa9−/− mice that were collected 7 days after cardiotoxin (CTX) injury. c, qPCR analysis of Hoxa9 expression in SCs isolated from tibialis anterior muscles injected with a self-delivering Hoxa9 or scrambled siRNA and collected 5 days after muscle injury. d, Representative immunofluorescence staining for Pax7 and laminin of injured tibialis anterior muscles from young adult and aged mice that were injected with a self-delivery siRNA and collected 7 days after muscle injury. Nuclei were counterstained with DAPI (blue). Arrowheads denote Pax7+ cells. e, Quantification of Pax7+ cells from d per area. f, Frequency distribution minimal Feret’s diameter of muscle fibres from d. g, Exemplary immunofluorescence staining for Pax7 and Ki67 on tibialis anterior muscles from aged Hoxa9+/+ and Hoxa9−/− mice collected 7 days after muscle injury. Nuclei were counterstained with DAPI (blue). h, Quantification of proliferating SCs (Ki67+/Pax7+) as depicted in g. Scale bars, 50 μm. P values were calculated by two-sided Student’s t-test (c, h) or two-way ANOVA (a, e, f). n = 3 mice in a; n = 3 mice in c; n = 3 mice in e, f; n = 4 mice in h.
Extended Data Figure 6 Inhibition of Hoxa9 improves regenerative capacity of aged SCs.
a, Quantification of donor-derived (eGFP+) myofibres from transplantation of SCs from young adult Hoxa9+/+ and Hoxa9−/− mice. b, qPCR analysis of Hoxa9 expression in SCs transduced with scrambled control or Hoxa9 shRNA encoding lentivirus. c–g, Transplantation of eGFP-labelled SCs from young adult and aged mice that were targeted with shRNAs against Hoxa9 or a scrambled control. c, Representative immunofluorescence staining for Pax7 and eGFP of transplanted muscle sections. Nuclei were counterstained with DAPI (blue). Arrowheads denote Pax7+/eGFP+ cells, asterisks label Pax7+/eGFP− cells. d, Quantification of donor-derived (eGFP+) Pax7+ cells in c. e, Representative immunofluorescence staining for eGFP and laminin of transplanted muscle sections, nuclei were counterstained with DAPI (blue). f, g, Quantification of donor-derived (eGFP+) myofibres in e for two different Hoxa9 shRNAs in two independent experiments. h, Exemplary immunofluorescence staining for eGFP and laminin in tibialis anterior muscles engrafted with untransduced aged SCs. Nuclei were counterstained with DAPI (blue). i, Flow cytometric analysis of transduction efficiency of donor SCs used for transplantation in primary recipients analysed in Fig. 2f. j, Representative flow cytometry plots for re-isolation of transplanted aged SCs that were untransduced as control or transduced with scrambled control or Hoxa9 shRNA encoding lentivirus as quantified in Fig. 2f. k, Representative immunofluorescence staining for eGFP and laminin in engrafted tibialis anterior muscles from secondary recipients quantified in Fig. 2g. Nuclei were counterstained with DAPI (blue). Scale bars, 20 μm (c), 50 μm (h) and 100 μm (e, k). P values were calculated by two-sided Student’s t-test (a, b) or two-way ANOVA (d, f, g). n = 4 recipient mice in a; n = 3 mice in b; n = 6 recipient mice (young donors), n = 4 recipient mice (aged donors) in d, f; n = 5 recipient mice in g.
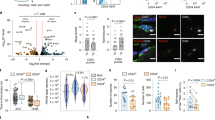
Extended Data Figure 7 Inhibition of Mll1 rescues H3K4me3 induction, Hoxa9 overexpression, and functional impairment of activated SCs from aged mice.
a, ChIP for H3K4me3 at promoters or exons of indicated Hox genes in activated SCs (4 day culture) from young adult and aged mice. b, Representative immunofluorescence staining for Pax7 and H3K4me3 on myofibre-associated SCs from aged mice that were freshly isolated or activated by 24-h culture of myofibres. c, Corrected total cell fluorescence (CTCF) for H3K4me3 on activated SCs shown in b. d, Representative immunofluorescence staining for Pax7 and Wdr5 on myofibre-associated SCs from young adult and aged mice that were freshly isolated or activated by 24-h culture of myofibres. e, CTCF for Mll1 and Wdr5 per activated SC as shown in d. f, qPCR analysis of Mll1 in SCs transfected with Mll1 siRNA or scrambled control. g, h, ChIPs for H3K4me3 (g) and Mll1 (h) in primary myoblasts 3 days after transfection with the indicated siRNAs. i, j, Immunofluorescence staining for Pax7 and Hoxa9 in myofibres from aged mice after transfection with Mll1 siRNA or scrambled control (i, quantification in Fig. 3d) or after treatment with OICR-9429 or vehicle (j). k, CTCF for Hoxa9 per SC as shown in j. l, Representative immunofluorescence staining for Pax7 and MyoD on OICR-9429 treated myofibre-associated SCs from aged mice after 72 h culture. Nuclei were counterstained with DAPI (blue). m, n, Average number of Pax7−/MyoD+ cells (m) or Pax7+/MyoD− cells (n) within clusters as shown in l. o, Representative immunofluorescence staining for Pax7 and MyoD on siRNA-treated myofibre-associated SCs from aged mice after 72-h culture. Nuclei were counterstained with DAPI (blue). p–r, Average number of Pax7−/MyoD+ cells (p), Pax7+/MyoD− cells (q) or Pax7+ cells (r) within clusters in o. s, Relative changes in cell number of aged SCs after treatment with OICR-9429 and 4 days of culture, compared to vehicle control. t, qPCR analysis of Mll1 in SCs transduced with Mll1 shRNA or scrambled control. u–w, Analysis of Pax7 expression in in vivo activated SCs from young adult and aged mice by RNA-sequencing (u), qPCR (v), or immunofluorescence as depicted in Fig. 1b (w). x, y, Pearson correlation comparing the Hoxa9 immunofluorescence signal (quantification in Fig. 1c) and the Pax7 immunofluorescence signal (quantification in w) of activated SCs from aged (x) and young adult (y) mice. Note, there is no correlation between Hoxa9 expression level and Pax7 expression level in activated SCs from aged mice. Scale bars, 20 μm (b, d, i, j, l, o). P values were calculated by two-way ANOVA (a, g, h), two-sided Student’s t-test (f, m, n, p–v), two-sided Mann–Whitney U-test (c, e, k, w) or Pearson correlation (x, y). n = 4 mice (young), n = 7 mice (aged) in a; n = 27 nuclei from 2 mice (young), n = 27 nuclei from 4 mice (aged) in c; n = 40/52 nuclei (Mll1), n = 44/99 nuclei (Wdr5) from 3 young/aged mice in e; n = 3 mice in f; n = 3 biological replicates (Wdr5 siRNA), n = 2 biological replicates (Mll1 siRNA) in g; n = 3 biological replicates in h; n = 173 nuclei (DMSO), n = 324 nuclei (OICR-9429) from 4 mice in k; n = 3 mice in m, n; n = 7 mice in p–r; n = 6 mice in s; n = 3 mice in t; n = 3 mice in u; n = 2 mice in v; n = 134 nuclei (young), n = 181 nuclei (aged) from 3 mice in w–y.
Extended Data Figure 8 Alterations in the epigenetic stress response of activated SCs from aged mice.
a, Heatmap displaying relative changes in abundance of different histone modifications (measured at the indicated peptides) in freshly isolated SCs from aged compared to young adult mice. SCs were analysed in quiescence (Q, derived from uninjured muscle) or at the indicated time points after activation mediated by muscle injury. Relative abundances at indicated days after injury are first normalized to quiescent SCs, and then compared between SCs isolated from aged and young adult mice and log2 scaled. Only significant changes are shown (P < 0.05). b, Expression analysis of the indicated genes in freshly isolated in vivo activated SCs from young adult and aged mice based on RNA-sequencing. c, Viability of primary myoblasts after 48-h treatment with bromodomain inhibitors (1 μM) from the Structural Genomics Consortium probe set, measured by Alamar Blue assay. d, Relative changes in cell number of aged SCs after treatment with non-toxic bromodomain inhibitors (1 μM) from c and 4 days of culture, compared to vehicle control. A Wilcoxon rank-sum test on the ratio of all cell counts being equal to 1 was performed to test the hypothesis of a general effect of the inhibitors on cell number. e, Representative immunofluorescence staining for Pax7 and Hoxa9 in siRNA-treated myofibre-associated SCs from aged mice. Scale bar, 20 μm. f, CTCF for Hoxa9 per SC as shown in e. g, Quantification of immunofluorescence staining for Hoxa9 in Pax7+ cells on myofibre-associated SCs from aged mice treated with bromodomain inhibitors. P values were calculated by two-sided Student’s t-test (a–c), Wilcoxon rank-sum test (d) or two-sided Mann–Whitney U-test (f, g). n = 4 mice in a; n = 3 mice in b; n = 4 biological replicates in c; n = 6 mice in d; n = 71 nuclei (scrambled siRNA), n = 48 nuclei (MOF siRNA), n = 98 nuclei (Utx siRNA) from 3 mice in f; n = 60 nuclei (vehicle), n = 59 nuclei (I-BRD9), n = 38 nuclei (LP99), n = 62 nuclei (PFI-3) from 3 mice in g.
Extended Data Figure 9 Overexpression of Hox genes inhibits SC function.
a, Expression of Hoxa9 in SCs transduced with Hoxa9 cDNA or eGFP as control. b, c, FACS-isolated SCs from young adult mice were transduced with a lentivirus either containing both eGFP and Hoxa9 cDNA or only eGFP. Infected (eGFP+) cells were isolated after 3 days. b, Frequency of myogenic colonies from single-cell-sorted SCs. c, Quantification of cell number based on Alamar Blue assay of bulk cultures. d, Frequency of myogenic colonies of SCs overexpressing the indicated Hox genes. e, g, TUNEL (e) or BrdU (g) staining of SCs overexpressing Hoxa9 or eGFP. Infected (eGFP+) cells were isolated 3 days after transduction and analysed 3 days later. Nuclei were counterstained with DAPI (blue). Arrowheads mark TUNEL- or BrdU-positive cells. f, h, Quantification of apoptosis (f) or proliferation (h) based on TUNEL or BrdU staining as in e or g. i, qPCR-based expression analysis of various cell-cycle and senescence markers in SCs overexpressing Hoxa9 compared to eGFP-infected controls, 5 days after infection. j, Senescence-associated-β-galactosidase (SA-β-Gal) staining of SCs overexpressing Hoxa9 or eGFP at day 5 after infection. Arrowheads mark SA-β-Gal-positive cells. k, Quantification of senescence per field of view (FOV) based on SA-β-Gal staining in j. l, Heatmap displaying log2 fold changes of expression of selected genes from microarray analysis in Fig. 5a. m–o, qPCR validation of differentially expressed genes annotated to Wnt (m), TGFβ (n) and JAK/STAT pathways (o) as in l. p, Identification of Hoxa9-binding sites by anti-HA ChIP of primary myoblasts overexpressing HA-tagged Hoxa9 cDNA or eGFP as control. Shown is the qPCR for 1 or 2 putative Hoxa9-binding sites at the indicated loci. Hoxa9-binding sites at target genes were identified as described in the Methods and are listed in Supplementary Table 1. A two-sided block bootstrap test on the difference of the percentage of bound DNA for all binding sites being equal to 0 was performed to test the hypothesis of a generally increased binding of Hoxa9. q–s, SCs were infected with lentiviruses expressing Hoxa9, Wnt3a, Bmp4 or Stat3 cDNAs or eGFP. qPCR analysis of expression of the indicated target genes at 5 days after infection: Axin2 (q), Bmp4 (r) and Stat3 (s). Scale bars, 20 μm (e, g) and 50 μm (j). P values were calculated by two-sided Student’s t-test (a–d, f, h, k, q–s) or two-way ANOVA (i, m–o). n = 4 mice in a; n = 3 mice in b; n = 7 mice in c; n = 3 mice in d; n = 4 mice in f, h, k; n = 3 mice (p15, p21), n = 6 mice (p16), n = 4 mice (all others) in i; n = 4 pools of 3 mice in l; n = 4 mice in m–o; n = 3 biological replicates for p; n = 3 mice (Wnt3a, Bmp4, Stat3), n = 4 mice (eGFP, Hoxa9) in q–s.
Extended Data Figure 10 Validation of Hoxa9 downstream targets.
a, Knockdown efficiency of two shRNAs (red bars) for Stat3, Bmp4 and Ctnnb1. b, SCs from young adult mice were transduced with an Hoxa9 and eGFP-encoding lentivirus. eGFP+ cells were sorted as single cells and cultured in the presence of noggin, DKK1 or 0.1% BSA in PBS as vehicle. Colony frequency was assessed after 5 days and is compared to Hoxa9 cDNA expressing cells treated with vehicle control. c, Representative immunofluorescence staining for Pax7 and MyoD on siRNA-transfected myofibres from aged mice after 72 h of culture. Nuclei were counterstained with DAPI (blue). d, e, Average number of Pax7+ cells (d) or Pax7−/MyoD+ cells (e) within clusters in c. f, Representative immunofluorescence staining for eGFP and laminin in tibialis anterior muscles engrafted with siRNA-transfected SCs isolated from eGFP transgenic aged mice. Nuclei were counterstained with DAPI (blue). g, Quantification of donor-derived (eGFP+) myofibres in f. h, Area-proportional Venn diagram of differentially expressed genes from indicated transcriptomes. i, Model for the Hoxa9-mediated impairment of SC function during ageing: quiescent SCs become activated upon muscle injury and proliferate as myoblasts to repair damaged muscle tissue. After activation, aged SCs display global and locus-specific alterations in the epigenetic stress response resulting in overexpression of Hoxa9, which in turn induces developmental pathways inhibiting SC function and muscle regeneration in aged mice. Scale bars, 20 μm (c), and 100 μm (f). P values were calculated by two-way ANOVA (a, b) or two-sided Student’s t-test (d, e, g). n = 3 mice in a; n = 4 mice in b; n = 5 mice in d, e; n = 5 recipient mice in g; n = 3 mice per group (activated SCs), n = 4 pools of 3 mice (Hoxa9 overexpression) in h.
Supplementary information
Supplementary Tables
This file contains Supplementary Table 1. (XLSX 29 kb)
Source data
Rights and permissions
About this article
Cite this article
Schwörer, S., Becker, F., Feller, C. et al. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432 (2016). https://doi.org/10.1038/nature20603
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20603
This article is cited by
-
Neurofibromin 1 controls metabolic balance and Notch-dependent quiescence of murine juvenile myogenic progenitors
Nature Communications (2024)
-
HOXB3 drives WNT-activation associated progression in castration-resistant prostate cancer
Cell Death & Disease (2023)
-
Chromatin accessibility dynamics of neurogenic niche cells reveal defects in neural stem cell adhesion and migration during aging
Nature Aging (2023)
-
Control of satellite cell function in muscle regeneration and its disruption in ageing
Nature Reviews Molecular Cell Biology (2022)
-
The landscape of aging
Science China Life Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.