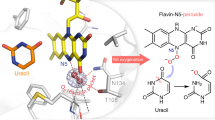
Abstract
The universal Per-ARNT-Sim (PAS) domain functions as a signal transduction module involved in sensing diverse stimuli such as small molecules, light, redox state and gases1,2. The highly evolvable PAS scaffold can bind a broad range of ligands, including haem, flavins and metal ions. However, although these ligands can support catalytic activity, to our knowledge no enzymatic PAS domain has been found. Here we report characterization of the first PAS enzyme: a haem-dependent oxidative N-demethylase. Unrelated to other amine oxidases, this enzyme contains haem, flavin mononucleotide, 2Fe-2S and tetrahydrofolic acid cofactors, and specifically catalyses the NADPH-dependent oxidation of dimethylamine. The structure of the α subunit reveals that it is a haem-binding PAS domain, similar in structure to PAS gas sensors3. The dimethylamine substrate forms part of a highly polarized oxygen-binding site, and directly assists oxygen activation by acting as both an electron and proton donor. Our data reveal that the ubiquitous PAS domain can make the transition from sensor to enzyme, suggesting that the PAS scaffold can support the development of artificial enzymes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Henry, J. T. & Crosson, S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65, 261–286 (2011)
Möglich, A., Ayers, R. A. & Moffat, K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294 (2009)
Gilles-Gonzalez, M. A. & Gonzalez, G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99, 1–22 (2005)
Philip, A. F., Kumauchi, M. & Hoff, W. D. Robustness and evolvability in the functional anatomy of a PER-ARNT-SIM (PAS) domain. Proc. Natl Acad. Sci. USA 107, 17986–17991 (2010)
Gardner, P. R. Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J. Inorg. Biochem. 99, 247–266 (2005)
Sun, S. et al. Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations. Arch. Biochem. Biophys. 545, 108–115 (2014)
Ward, T. R. Artificial metalloenzymes based on the biotin-avidin technology: enantioselective catalysis and beyond. Acc. Chem. Res. 44, 47–57 (2011)
Alberta, J. A. & Dawson, J. H. Purification to homogeneity and initial physical characterization of secondary amine monooxygenase. J. Biol. Chem. 262, 11857–11863 (1987)
Alberta, J. A., Andersson, L. A. & Dawson, J. H. Spectroscopic characterization of secondary amine mono-oxygenase. Comparison to cytochrome P-450 and myoglobin. J. Biol. Chem. 264, 20467–20473 (1989)
Chistoserdova, L. Modularity of methylotrophy, revisited. Environ. Microbiol. 13, 2603–2622 (2011)
Colby, J. & Zatman, L. J. Enzymological aspects of the pathways for trimethylamine oxidation and C1 assimilation of obligate methylotrophs and restricted facultative methylotrophs. Biochem. J. 148, 513–520 (1975)
Guengerich, F. P. & Munro, A. W. Unusual cytochrome p450 enzymes and reactions. J. Biol. Chem. 288, 17065–17073 (2013)
Dziewit, L. et al. Genome-guided insight into the methylotrophy of Paracoccus aminophilus JCM 7686. Front. Microbiol. 6, 852 (2015)
Tralau, T. et al. An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde. J. Biol. Chem. 284, 17826–17834 (2009)
Singh, H., Arentson, B. W., Becker, D. F. & Tanner, J. J. Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc. Natl Acad. Sci. USA 111, 3389–3394 (2014)
LoBrutto, R., Wei, Y.-H., Mascarenhas, R., Scholes, C. P. & King, T. E. Electron nuclear double resonance and electron paramagnetic resonance study on the structure of the NO-ligated heme alpha 3 in cytochrome c oxidase. J. Biol. Chem. 258, 7437–7448 (1983)
Morse, R. H. & Chan, S. I. Electron paramagnetic resonance studies of nitrosyl ferrous heme complexes. Determination of an equilibrium between two conformations. J. Biol. Chem. 255, 7876–7882 (1980)
Sawai, H. et al. Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem. Commun. (Camb.) 48, 6523–6525 (2012)
Sono, M., Roach, M. P., Coulter, E. D. & Dawson, J. H. Heme-Containing Oxygenases. Chem. Rev. 96, 2841–2888 (1996)
Matsunaga, I. & Shiro, Y. Peroxide-utilizing biocatalysts: structural and functional diversity of heme-containing enzymes. Curr. Opin. Chem. Biol. 8, 127–132 (2004)
Rittle, J. & Green, M. T. Cytochrome P450 compound I: capture, characterization, and C-H bond activation kinetics. Science 330, 933–937 (2010)
Daiber, A. et al. Isotope effects and intermediates in the reduction of NO by P450(NOR) . J. Inorg. Biochem. 88, 343–352 (2002)
Hofrichter, M. & Ullrich, R. Heme-thiolate haloperoxidases: versatile biocatalysts with biotechnological and environmental significance. Appl. Microbiol. Biotechnol. 71, 276–288 (2006)
Roberts, A. G. et al. NMR-derived models of amidopyrine and its metabolites in complexes with rabbit cytochrome P450 2B4 reveal a structural mechanism of sequential N-dealkylation. Biochemistry 50, 2123–2134 (2011)
Du, Y. et al. Conversion of a heme-based oxygen sensor to a heme oxygenase by hydrogen sulfide: effects of mutations in the heme distal side of a heme-based oxygen sensor phosphodiesterase (Ec DOS). Biometals 26, 839–852 (2013)
Fitzpatrick, P. F. Oxidation of amines by flavoproteins. Arch. Biochem. Biophys. 493, 13–25 (2010)
Brazeau, B. J., Johnson, B. J. & Wilmot, C. M. Copper-containing amine oxidases. Biogenesis and catalysis; a structural perspective. Arch. Biochem. Biophys. 428, 22–31 (2004)
Walport, L. J., Hopkinson, R. J. & Schofield, C. J. Mechanisms of human histone and nucleic acid demethylases. Curr. Opin. Chem. Biol. 16, 525–534 (2012)
Green, J. & Large, P. J. Oxidation of dimethylamine and trimethylamine in methazotrophic yeasts by microsomal mono-oxygenases sensitive to carbon monoxide. Biochem. Biophys. Res. Commun. 113, 900–907 (1983)
Bertrand, P., More, C. & Camensuli, P. Evidence for a magic magnetic configuration between FMN and the [2Fe-2S]+ center of phthalate dioxygenase reductase of Pseudomonas cepacia. J. Am. Chem. Soc. 117, 1807–1809 (1995)
Berry, E. A. & Trumpower, B. L. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 (1987)
Morrison, J. F. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta 185, 269–286 (1969)
Zurek, G. & Karst, U. Microplate photometric determination of aldehydes in disinfectant solutions. Anal. Chim. Acta 351, 247–257 (1997)
Nash, T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem. J. 55, 416–421 (1953)
Dutton, P. L. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 54, 411–435 (1978)
Munro, A. W., Noble, M. A., Robledo, L., Daff, S. N. & Chapman, S. K. Determination of the redox properties of human NADPH-cytochrome P450 reductase. Biochemistry 40, 1956–1963 (2001)
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011)
Acknowledgements
This work was supported by BBSRC grant (BBE0170101). We thank the BBSRC/EPSRC SYNBIOCHEM Centre (grant BB/M017702/1) for access to analytical equipment. We thank the Diamond Light Source for access to beamlines (proposal number MX8997). D.L. is a Royal Society Wolfson Merit Award holder. N.S.S. is an EPSRC Established Career Fellow. The authors acknowledge the use of the Computational Shared Facility and the Protein Structure Facility at The University of Manchester.
Author information
Authors and Affiliations
Contributions
M.O. carried out molecular biology, biophysical and structural biology studies on the P. mendocina HODM. M.O. and L.D. carried out purification and characterisation of variant haem domain forms. P.L., B.M. and T.T. were involved in initial screening and solution characterization of HODM homologues. T.T. carried out in vivo formaldehyde detection. K.F. and S.E.J.R. performed and analysed EPR experiments. S.H. performed DFT calculations and kinetic data analysis. All authors discussed the results and participated in writing the manuscript. D.L. initiated and directed this research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks P. Ortiz de Montellano, C. Wilmot and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Catabolic pathway featuring HODM and additional HODM characterization in vivo and in vitro.
a, Trimethylamine degradation pathway with links to C1 metabolism in proteobacteria. 1, trimethylamine monooxygenase; 2, trimethylamine N-oxide demethylase; 3, haem-dependent oxidative demethylase (HODM), also known as secondary amine monooxygenase (SAMO); 4, methylamine dehydrogenase (EC 1.4.99.3); 5, methylamine oxidase (EC 1.4.3.21); 6, N-methylglutamate synthase (EC 2.1.1.21); 7, N-methylglutamate dehydrogenase (EC 1.5.99.5); 8, γ-glutamylmethylamide synthetase (EC 6.3.4.12); 9, γ- glutamylmethylamide-dissimilating enzyme; 10, methylene-H4F dehydrogenase/cyclohydrolase (EC 1.5.1.15); 11, formyl-H4F deformylase/ formyl-H4F synthetase (EC 3.5.1.10); 12, formate dehydrogenase (EC 1.2.1.2). Formaldehyde can be assimilated via the ribulose monophosphate (RuMP) pathway or, via methylene-tetrahydrofolate, through formation of serine. The inset shows an SDS–PAGE gel of purified recombinant HODM, revealing bands corresponding to all four subunits. Additional bands are visible that appear to correspond to multimeric forms or proteolytic fragments of HODM subunits as verified by mass spectrometry of tryptic digests. b, Formaldehyde leakage from HODM in vivo. Efficient detoxification of formaldehyde requires the γ subunit as well as tetrahydrofolic acid (THF). Cells containing wild-type and mutant HODM enzyme (Δγ, deletion of subunit gamma) from Methylobacillus flagellatus (strain KT) were grown in the presence of DMA (grey bars) or DMA and glycine (shaded bars), respectively. Fluorescence readings were subsequently corrected for background activity and scaled with the mutant enzyme set as 100%. The data show that glycine, as well as the γ subunit, have strong effects on enzymatic formaldehyde production. While the first increases intracellular levels of THF, the latter is required for THF binding. c, The HODM ferrous-oxy complex is long-lived at room temperature. The ferrous-oxy decay to the ferric state was monitored at 409 nm and plotted as a function of time. The decay curve observed was fitted using the exponential decay equation to derive a half-life of 50 ± 10 min with a decay rate of k = 0.014 ± 0.001 min−1. d, Determination of the redox potential of the HODM haem subunit. The main panel shows UV-visible spectra for the HODM haem domain during a redox titration. The oxidized enzyme (thick black line, spectrum recorded at −76 mV versus normal hydrogen electrode) has its Soret maximum at 409 nm. The fully reduced enzyme (thick line, spectrum recorded at −317 mV versus NHE) has its Soret maximum at 422 nm. Intermediate spectra are indicated in thin lines, and there is an isosbestic point at approximately 415 nm. The HODM haem domain also displays increased absorbance in the Q-band region (~520–620 nm) in the reduced (ferrous) state. The inset shows a plot of absorbance change (ΔA422 minus ΔA409) versus applied potential (versus NHE) fitted using the Nernst equation. This provides a midpoint reduction potential value for the HDOM haem domain FeIII/FeII couple of 128 ± 4 mV versus NHE. e, f, DMA binding to ferrous HODM (e) and DMA binding to ferrous-oxy HODM (f). Absorbance changes (as shown in the inset) as a function of DMA concentration are fitted using a hyperbolic function/Morrison equation, leading to a Kd = 1.7 ± 0.2 mM for ferrous HODM (ferric HODM has a Kd of 1.5 ± 0.1 mM, data not shown) and a Kd = 15 ± 3 μM for ferrous-oxy HODM (errors bars are s.e.m., n = 3).
Extended Data Figure 2 X-band continuous-wave electron paramagnetic resonance spectra of anaerobically purified HODM.
a, Anaerobically purified HODM exposed to air for two minutes. b, The HODM enzyme shown in a with the addition of excess DMA. c, Anaerobically purified HODM. d, The HODM enzyme shown in c with the addition of excess DMA. Experimental parameters: microwave power, 0.5 mW; modulation amplitude, 3 G; temperature, 30 K. e, HODM alone. f, HODM plus excess DMA. g, HODM plus excess diethylamine. h, HODM plus excess piperidine. Experimental parameters: microwave power, 0.5 mW; modulation amplitude, 3 G; temperature, 30 K. Only DMA (f) shows the loss of the 14N His superhyperfine coupling in the ferrohaem–NO component of the spectrum and a significant change in the g values exhibited by the [2Fe-2S]1+ component of the spectrum.
Extended Data Figure 3 HODM cannot make use of peroxide.
Spectral changes observed following mixing of 10 μM ferric HODM with 200 μM H2O2. The haem Soret peak (black line) decays upon H2O2 addition (200 μM) (thin grey lines each after 30 s) with further decay at 5 min incubation (thin black line) and 60 min (black dotted line). The inset shows a similar experiment conducted in the presence of 2 mM DMA.
Extended Data Figure 4 The HODM haem subunit has similar properties to the HODM holoenzyme.
a, UV-visible absorption features of alpha HODM (8.5 μM). Ferric (thick solid line), ferrous (thin solid line) Fe2+–O2 (dotted line) and Fe2+–CO (dashed line). The major (Soret) absorption band is centred at 409, 423, 418 and 421 nm, respectively. The α/β bands are magnified in the inset. b, Titration of ferric and ferrous-oxy HODM α subunit with DMA. Absorbance changes associated with the haem Soret peak are plotted as a function of DMA concentration. The data are fitted using a hyperbolic function or the Morrison equation. Top, ferric with Kd = 12.6 ± 0.2 mM. Bottom, ferrous-oxy with Kd = 41.0 ± 5.0 μM (errors bars are s.e.m., n = 3). Insets show UV-vis spectra of the titration of DMA (0–50 mM) against ferric and ferrous-oxy HODM α subunit (7.5/4.8 μM). The UV-vis spectrum of α in buffer A was recorded initially and then after each addition of DMA. The direction of the absorbance changes of the ferric and ferrous-oxy Soret peak on substrate addition is measured from 409/415 nm (thick line) to 416/414 nm (dotted line), respectively. c–e, Spectral properties of HODM haem subunit variants E266Q, W180A and R224A, respectively. Ferric (thick black line), ferric in presence of 1 mM DMA (thick grey line), ferrous (thin black line), ferrous in presence of 1 mM DMA (thin grey line) and Fe2+–O2 (dotted line). The α/β bands are magnified in the inset. While the E226Q variant lacks spectroscopic features associated with DMA binding, the R224A variant appears to bind DMA as a 6th ligand to the haem iron in the ferrous state. Oxygen binding can be detected for all variants.
Extended Data Figure 5 Possible routes for DMA oxidation in HODM.
Following formation of compound 0, various possibilities exist for O–O bond cleavage in principle. A direct hydride transfer from the substrate is shown in A, bypassing the need for higher oxidation states of the haem Fe. Route B depicts the homolytic bond cleavage with formation of compound II and an amine radical. Route C depicts a P450-like mechanism, with formation of the highly oxidizing compound I. Both the B and C routes can give rise to various products. The N-oxide product is not observed with DMA as a substrate, which results in stoichiometric production of formaldehyde in vitro. This can be derived from either the iminium or hydroxylamine products.
Extended Data Figure 6 X-ray-induced photoreduction of the HODM haem subunit in complex with DMA and O2.
The initial spectrum recorded before X-ray illumination (ESRF beamline ID14-1 (wavelength 0.934 Å)) was subtracted from the post illumination spectrum. A shift in the α/β bands is recorded with a decrease at 575 nm and an increase at 556 nm. This corresponds to formation of the ferrous species (merged α/β band at 556 nm).
Extended Data Figure 7 MA-dependent NADPH oxidation by HODM in absence of formaldehyde production.
The kinetic curve (mean values of initial rate plotted as a function of MA concentration) was fitted using the Michaelis–Menten function kcat = 7.0 ± 0.1 s−1 and KM = 0.55 ± 0.04 mM. Inset shows Purpald colourimetric assay for detection of substrate-dependent formaldehyde production. Data are represented as a percentage of expected formaldehyde produced for 1:1 conversion of DMA/MA to formaldehyde. Error bars represent s.d.
Extended Data Figure 8 Gas phase density functional theory and ab initio comparison of the energies of DMA- and MA-based reaction intermediates.
Quoted energies in the table are the sum of electronic and thermal enthalpies for structures in the gas phase at 298.15 K with units of hartrees. Values in parentheses are the difference relative to the neutral amine. The relative energies of the DMA species are lower than those of the MA species by 10–65 kJ mol−1 (0.04–0.025 hartrees). The pKa valuesof DMA (R = CH3) and MA (R = H) (1) are very similar (10.64 and 10.62 respectively). Calculations were performed at both the (U)B3LYP/6-311++G(d,p) and (U)MP2/6-311++G(d,p) level using the ‘Freq’ keyword in Gaussian 09 rev. B.01.
Extended Data Figure 9 Multiple sequence alignment of HODM including Aer2 PAS domain secondary structure information.
Alignment of HODM proteins from the DUF3445 family from the bacteria: Pseudomonas mendocina (Pmen), Rhizobium etli (Retl), Candidatus Puniceispirillum marinum (Cpun) and Gordonia bronchialis (Gbro), and from the fungi Candida dubliniensis (Cdub) and Nectria haematococca (Nhae). The catalytic Arg, Glu and Gln residues and the substrate-discriminating Trp are marked with black dots. The conserved haem-binding His in Pmen and Aer2 are marked with a grey and a black asterisk, respectively. Secondary structural elements are highlighted, PAS-domain-specific α-helices in blue and β-sheets in pink, and HODM-specific α-helices in pale grey and β-sheets in dark grey.
Rights and permissions
About this article
Cite this article
Ortmayer, M., Lafite, P., Menon, B. et al. An oxidative N-demethylase reveals PAS transition from ubiquitous sensor to enzyme. Nature 539, 593–597 (2016). https://doi.org/10.1038/nature20159
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20159
This article is cited by
-
Genomic profiling and characteristics of a C1 degrading heterotrophic fresh-water bacterium Paracoccus sp. strain DMF
Archives of Microbiology (2024)
-
Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein
Nature Chemical Biology (2018)
-
Evolution of chalcone isomerase from a noncatalytic ancestor
Nature Chemical Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.