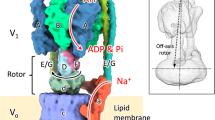
Abstract
Vacuolar-type ATPases (V-ATPases) are ATP-powered proton pumps involved in processes such as endocytosis, lysosomal degradation, secondary transport, TOR signalling, and osteoclast and kidney function. ATP hydrolysis in the soluble catalytic V1 region drives proton translocation through the membrane-embedded VO region via rotation of a rotor subcomplex. Variability in the structure of the intact enzyme has prevented construction of an atomic model for the membrane-embedded motor of any rotary ATPase1,2,3,4,5. We induced dissociation and auto-inhibition of the V1 and VO regions of the V-ATPase by starving the yeast Saccharomyces cerevisiae6,7, allowing us to obtain a ~3.9-Å resolution electron cryomicroscopy map of the VO complex and build atomic models for the majority of its subunits. The analysis reveals the structures of subunits ac8c′c″de and a protein that we identify and propose to be a new subunit (subunit f). A large cavity between subunit a and the c-ring creates a cytoplasmic half-channel for protons. The c-ring has an asymmetric distribution of proton-carrying Glu residues, with the Glu residue of subunit c″ interacting with Arg735 of subunit a. The structure suggests sequential protonation and deprotonation of the c-ring, with ATP-hydrolysis-driven rotation causing protonation of a Glu residue at the cytoplasmic half-channel and subsequent deprotonation of a Glu residue at a luminal half-channel.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Zhao, J., Benlekbir, S. & Rubinstein, J. L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 521, 241–245 (2015)
Allegretti, M. et al. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature 521, 237–240 (2015)
Zhou, A. et al. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 4, e10180 (2015)
Morales-Rios, E., Montgomery, M. G., Leslie, A. G. W. & Walker, J. E. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc. Natl Acad. Sci. USA 112, 13231–13236 (2015)
Schep, D. G., Zhao, J. & Rubinstein, J. L. Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance. Proc. Natl Acad. Sci. USA 113, 3245–3250 (2016)
Kane, P. M. Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem . 270, 17025–17032 (1995)
Sumner, J. P. et al. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J. Biol. Chem. 270, 5649–5653 (1995)
Couoh-Cardel, S., Milgrom, E. & Wilkens, S. Affinity purification and structural features of the yeast vacuolar ATPase Vo membrane sector. J. Biol. Chem. 290, 27959–27971 (2015)
Benlekbir, S., Bueler, S. A. & Rubinstein, J. L. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol. 19, 1356–1362 (2012)
Qi, J. & Forgac, M. Function and subunit interactions of the N-terminal domain of subunit a (Vph1p) of the yeast V-ATPase. J. Biol. Chem. 283, 19274–19282 (2008)
Powell, B., Graham, L. A. & Stevens, T. H. Molecular characterization of the yeast vacuolar H+-ATPase proton pore. J. Biol. Chem. 275, 23654–23660 (2000)
Hirata, R., Graham, L. A., Takatsuki, A., Stevens, T. H. & Anraku, Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J. Biol. Chem. 272, 4795–4803 (1997)
Nishi, T., Kawasaki-Nishi, S. & Forgac, M. The first putative transmembrane segment of subunit c" (Vma16p) of the yeast V-ATPase is not necessary for function. J. Biol. Chem. 278, 5821–5827 (2003)
Couoh-Cardel, S., Hsueh, Y.-C., Wilkens, S. & Movileanu, L. Yeast V-ATPase proteolipid ring acts as a large-conductance transmembrane protein pore. Sci. Rep. 6, 24774 (2016)
Matthies, D. et al. High-resolution structure and mechanism of an F/V-hybrid rotor ring in a Na+-coupled ATP synthase. Nat. Commun. 5, 5286 (2014)
Vik, S. B. & Antonio, B. J. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 269, 30364–30369 (1994)
Junge, W., Lill, H. & Engelbrecht, S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22, 420–423 (1997)
Neale, C., Chakrabarti, N., Pomorski, P., Pai, E. F. & Pomès, R. Hydrophobic gating of ion permeation in magnesium channel CorA. PLOS Comput. Biol. 11, e1004303 (2015)
Angevine, C. M., Herold, K. A. & Fillingame, R. H. Aqueous access pathways in subunit a of rotary ATP synthase extend to both sides of the membrane. Proc. Natl Acad. Sci. USA 100, 13179–13183 (2003)
Toei, M., Toei, S. & Forgac, M. Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J. Biol. Chem. 286, 35176–35186 (2011)
Kühlbrandt, W. & Davies, K. M. Rotary ATPases: a new twist to an ancient machine. Trends Biochem. Sci. 41, 106–116 (2016)
Pellegrini-Calace, M., Maiwald, T. & Thornton, J. M. PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLOS Comput. Biol. 5, e1000440 (2009)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996)
Kawasaki-Nishi, S., Nishi, T. & Forgac, M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc. Natl Acad. Sci. USA 98, 12397–12402 (2001)
Cain, B. D. & Simoni, R. D. Impaired proton conductivity resulting from mutations in the a subunit of F1F0 ATPase in Escherichia coli. J. Biol. Chem. 261, 10043–10050 (1986)
DeCoursey, T. E. The voltage-gated proton channel: a riddle, wrapped in a mystery, inside an enigma. Biochemistry 54, 3250–3268 (2015)
Bueler, S. A. & Rubinstein, J. L. Vma9p need not be associated with the yeast V-ATPase for fully-coupled proton pumping activity in vitro. Biochemistry 54, 853–858 (2015)
Nelson, H. & Nelson, N. Disruption of genes encoding subunits of yeast vacuolar H+-ATPase causes conditional lethality. Proc. Natl Acad. Sci. USA 87, 3503–3507 (1990)
Compton, M. A., Graham, L. A. & Stevens, T. H. Vma9p (subunit e) is an integral membrane VO subunit of the yeast V-ATPase. J. Biol. Chem. 281, 15312–15319 (2006)
MacCallum, J. L., Bennett, W. F. D. & Tieleman, D. P. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 94, 3393–3404 (2008)
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996)
Olsen, J. V. et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 (2005)
Marr, C. R., Benlekbir, S. & Rubinstein, J. L. Fabrication of carbon films with ∼ 500nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014)
Tivol, W. F., Briegel, A. & Jensen, G. J. An improved cryogen for plunge freezing. Microsc. Microanal. 14, 375–379 (2008)
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007)
Rubinstein, J. L. & Brubaker, M. A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 192, 188–195 (2015)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Zhao, J., Brubaker, M. A., Benlekbir, S. & Rubinstein, J. L. Description and comparison of algorithms for correcting anisotropic magnification in cryo-EM images. J. Struct. Biol. 192, 209–215 (2015)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005)
Grant, T. & Grigorieff, N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J. Struct. Biol. 192, 204–208 (2015)
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015)
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015)
Grigorieff, N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 (2007)
Grigorieff, N. Frealign: an exploratory tool for single-particle Cryo-EM. Methods Enzymol . 579, 191–226 (2016)
Penczek, P. A. et al. CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy 140, 9–19 (2014)
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013)
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr . 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr . 66, 213–221 (2010)
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015)
Acknowledgements
We thank N. Grigorieff for providing access to the Titan Krios electron microscope and H. Urlaub for providing C.S. with access to mass spectrometry instrumentation while in Göttingen. We thank Z. Ripstein, P. Tieleman, R. Pomès, and J.-P. Julien for discussions and J. Zhao, J.-P. Julien and V. Kanelis for a critical reading of the manuscript. M.T.M.-J. was supported by a Postdoctoral Fellowship from the Canadian Institutes of Health Research (CIHR), C.V.R. is a Royal Society Professor and J.L.R. holds a Canada Research Chair. This work was supported by operating grant MOP81294 from the Canadian Institutes of Health Research (J.L.R.), Wellcome Trust grants WT008150 and WT099141 (C.V.R.), and European Research Council IMPRESS grant ERC268851 (C.V.R.).
Author information
Authors and Affiliations
Contributions
M.T.M.-J. prepared yeast strains, purified protein, prepared cryo-EM specimens, performed 200 kV cryo-EM and Relion image analysis, and built the atomic models. A.R. collected and pre-processed data with the 300-kV microscope and performed image analysis with Frealign. C.S. performed the mass spectrometry analysis. S.A.B. prepared yeast strains and assisted with protein purification. S.B. assisted with cryo-EM specimen preparation and screening. C.V.R. supervised mass spectrometry experiments, and J.L.R. supervised the other aspects of the work and coordinated experiments. J.L.R. and M.T.M.-J. wrote the manuscript and prepared the figures with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks A. Hack, K. Parra, H. Saibil and J. Weber for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Subunit composition of the intact V-ATPase and dissociated V1 and VO regions.
The rotor is outlined in black and the two half-channels in the VO region are indicated with dashed lines. The intact V-ATPase (left) dissociates into the auto-inhibited V1 and VO complexes upon nutrient starvation. Figure adapted from ref. 1.
Extended Data Figure 2 Cryo-EM map generation.
a, An example micrograph with protein particles circled in red. Scale bar, 500 Å. b, Fourier shell correlation (FSC) curve. The highest-resolution information used in image alignment (6 Å) and the overall resolution of the map at FSC = 0.143 (3.9 Å) are indicated. c, Local resolution assessment. Scale bar, 25 Å. d, Image orientation distribution. e, Example 2D class average images.
Extended Data Figure 3 Model building.
a–f, Example regions of the atomic model built for subunits a (a), c″ and c′ (b), c(1) (c), d (d), e (e), and f (f). g, The different α-helices from the c-ring bearing conserved Glu residues show variable resolution. An α-helix from the N-terminal domain of subunit a has poor resolution. Residue numbers are shown in brackets.
Extended Data Figure 4 VO complex lacking subunit d.
a, The VO complex map from all of the particle images shows subunit d. b, VO complex map from a 3D class, containing 24,744 particle images, that lacks subunit d was determined at 7.8-Å resolution. Scale bar, 25 Å.
Extended Data Figure 5 VO complex is in rotational state 3
. a–c, rotational states 1, 2, and 3 of the intact V-ATPase show the two α helices of subunit c" within the c-ring1. d, The two α-helices of subunit c″ within the c-ring show the ring to be in the same orientation as in rotational state 3 of the intact V-ATPase. Scale bar, 25 Å.
Extended Data Figure 6 Identification of subunit f.
a, SDS–PAGE gel (left) and western blot (right) against a 3×FLAG-tag for the affinity purification of 3×FLAG-tagged YPR170W-B (subunit f) and Vma1p (subunit A) show that both proteins are components of the V-ATPase. b, Surface-rendered 3D maps (upper) and map cross-sections (lower) showing the wild-type VO complex (left) and the VO complex from a yeast strain with the YPR170W-B gene deleted (right). Density from YPR170W-B is indicated with a red arrow. Scale bar, 25 Å. c, Yeast strains with the STV1 and VPH1 genes deleted, the STV1 and YPR170W-B gene deleted, and only STV1 gene deleted were grown on both YPD medium (left) and YPD medium with zinc (right), demonstrating that deletion of YPR170W-B does not cause the VMA phenotype.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-2. Table 1 contains data acquisition, processing, and model statistics and Table 2 contains a summary of the mass spectrometry results for candidate proteins identified in the membrane region of the V-ATPase. (PDF 602 kb)
Supplementary Data
This file contains a spreadsheet showing the mass spectrometry database search results. (XLSX 32 kb)
Rights and permissions
About this article
Cite this article
Mazhab-Jafari, M., Rohou, A., Schmidt, C. et al. Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature 539, 118–122 (2016). https://doi.org/10.1038/nature19828
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19828
This article is cited by
-
The a subunit isoforms of vacuolar-type proton ATPase exhibit differential distribution in mouse perigastrulation embryos
Scientific Reports (2022)
-
The V-ATPases in cancer and cell death
Cancer Gene Therapy (2022)
-
Drosophila melanogaster: a simple genetic model of kidney structure, function and disease
Nature Reviews Nephrology (2022)
-
Coordinated conformational changes in the V1 complex during V-ATPase reversible dissociation
Nature Structural & Molecular Biology (2022)
-
Vacuolar-type proton ATPase is required for maintenance of apicobasal polarity of embryonic visceral endoderm
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.