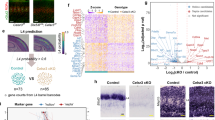
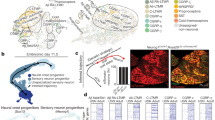
Abstract
Modality-specific sensory inputs from individual sense organs are processed in parallel in distinct areas of the neocortex. For each sensory modality, input follows a cortico–thalamo–cortical loop in which a ‘first-order’ exteroceptive thalamic nucleus sends peripheral input to the primary sensory cortex, which projects back to a ‘higher order’ thalamic nucleus that targets a secondary sensory cortex1,2,3,4,5,6. This conserved circuit motif raises the possibility that shared genetic programs exist across sensory modalities. Here we report that, despite their association with distinct sensory modalities, first-order nuclei in mice are genetically homologous across somatosensory, visual, and auditory pathways, as are higher order nuclei. We further reveal peripheral input-dependent control over the transcriptional identity and connectivity of first-order nuclei by showing that input ablation leads to induction of higher-order-type transcriptional programs and rewiring of higher-order-directed descending cortical input to deprived first-order nuclei. These findings uncover an input-dependent genetic logic for the design and plasticity of sensory pathways, in which conserved developmental programs lead to conserved circuit motifs across sensory modalities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Guillery, R. W. Anatomical evidence concerning the role of the thalamus in corticocortical communication: a brief review. J. Anat. 187, 583–592 (1995)
Sherman, S. M. S. Thalamocortical interactions. Curr. Opin. Neurobiol. 22, 575–579 (2012)
Herkenham, M. Laminar organization of thalamic projections to the rat neocortex. Science 207, 532–535 (1980)
Smith, P. H., Uhlrich, D. J., Manning, K. A. & Banks, M. I. Thalamocortical projections to rat auditory cortex from the ventral and dorsal divisions of the medial geniculate nucleus. J. Comp. Neurol. 520, 34–51 (2012)
Clascá, F., Rubio-Garrido, P. & Jabaudon, D. Unveiling the diversity of thalamocortical neuron subtypes. Eur. J. Neurosci. 35, 1524–1532 (2012)
Theyel, B. B., Llano, D. A. & Sherman, S. M. The corticothalamocortical circuit drives higher-order cortex in the mouse. Nat. Neurosci. 13, 84–88 (2010)
Pouchelon, G. et al. Modality-specific thalamocortical inputs instruct the identity of postsynaptic L4 neurons. Nature 511, 471–474 (2014)
Frangeul, L. et al. Specific activation of the paralemniscal pathway during nociception. Eur. J. Neurosci. 39, 1455–1464 (2014)
Viaene, A. N., Petrof, I. & Sherman, S. M. Properties of the thalamic projection from the posterior medial nucleus to primary and secondary somatosensory cortices in the mouse. Proc. Natl Acad. Sci. USA 108, 18156–18161 (2011)
Chou, S.-J. et al. Geniculocortical input drives genetic distinctions between primary and higher-order visual areas. Science 340, 1239–1242 (2013)
Wang, S., Eisenback, M. A. & Bickford, M. E. Relative distribution of synapses in the pulvinar nucleus of the cat: implications regarding the “driver/modulator” theory of thalamic function. J. Comp. Neurol. 454, 482–494 (2002)
Reichova, I. & Sherman, S. M. Somatosensory corticothalamic projections: distinguishing drivers from modulators. J. Neurophysiol. 92, 2185–2197 (2004)
Lee, C. C. & Sherman, S. M. Synaptic properties of thalamic and intracortical inputs to layer 4 of the first- and higher-order cortical areas in the auditory and somatosensory systems. J. Neurophysiol. 100, 317–326 (2008)
Grant, E., Hoerder-Suabedissen, A. & Molnár, Z. The regulation of corticofugal fiber targeting by retinal inputs. Cereb. Cortex 26, 1336–1348 (2016)
Telley, L. et al. Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446 (2016)
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015)
Lu, E., Llano, D. A. & Sherman, S. M. Different distributions of calbindin and calretinin immunostaining across the medial and dorsal divisions of the mouse medial geniculate body. Hear. Res. 257, 16–23 (2009)
Shanks, J. A. et al. Corticothalamic axons are essential for retinal ganglion cell axon targeting to the mouse dorsal lateral geniculate nucleus. J. Neurosci. 36, 5252–5263 (2016)
Gong, S. et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci. 27, 9817–9823 (2007)
Butler, A. B. Evolution of the thalamus: a morphological and functional review. Thalamus Relat. Syst. 4, 1–24 (2008)
Bishop, G. H. The relation between nerve fiber size and sensory modality: phylogenetic implications of the afferent innervation of cortex. J. Nerv. Ment. Dis. 128, 89–114 (1959)
Wong-Riley, M. T. & Welt, C. Histochemical changes in cytochrome oxidase of cortical barrels after vibrissal removal in neonatal and adult mice. Proc. Natl Acad. Sci. USA 77, 2333–2337 (1980)
Wong-Riley, M. Changes in the visual system of monocularly sutured or enucleated cats demonstrable with cytochrome oxidase histochemistry. Brain Res. 171, 11–28 (1979)
Yamashita, T. et al. Membrane potential dynamics of neocortical projection neurons driving target-specific signals. Neuron 80, 1477–1490 (2013)
Bavelier, D. & Neville, H. J. Cross-modal plasticity: where and how? Nat. Rev. Neurosci. 3, 443–452 (2002)
De la Rossa, A. & Jabaudon, D. In vivo rapid gene delivery into postmitotic neocortical neurons using iontoporation. Nat. Protocols 10, 25–32 (2015)
Golding, B. et al. Retinal input directs the recruitment of inhibitory interneurons into thalamic visual circuits. Neuron 81, 1057–1069 (2014)
Scholkopf, B., Smola, A. J., Williamson, R. C. & Bartlett, P. L. New support vector algorithms. Neural Comput. 12, 1207–1245 (2000)
Noble, W. S. What is a support vector machine? Nat. Biotechnol. 24, 1565–1567 (2006)
Guyon, I., Weston, J., Barnhill, S. & Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 46, 389–422 (2002)
Magdaleno, S. et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 4, e86 (2006)
Bian, W.-J., Miao, W.-Y., He, S.-J., Qiu, Z. & Yu, X. Coordinated spine pruning and maturation mediated by inter-spine competition for cadherin/catenin complexes. Cell 162, 808–822 (2015)
Acknowledgements
We thank A. Benoit, S. Binvignat, M. Lanzillo and members of the Genomics Platform of the University of Geneva for technical assistance, A. Holtmaat and A. Carleton for the gift of the transgenic mouse lines. We thank J. Prados for help in the bioinformatics analysis. We thank E. Azim, A. Holtmaat, M. Scanziani and S. Tole for helpful comments on the manuscript. Work in the Jabaudon laboratory is supported by the Swiss National Science Foundation (SNF) (PP00P3_123447), the Leenaards Foundation, the Synapsis Foundation and the NARSAD Foundation.
Author information
Authors and Affiliations
Contributions
G.P., L.F. and D.J. conceived the project; G.P., L.F. and S.L. performed the experiments; L.T., G.P., L.F., S.L. and D.J. performed analyses; D.J. and L.F. wrote the manuscript with help of all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks S. Nelson, F. Polleux and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Hierarchical order is the primary determinant of transcriptional identity in somatosensory and visual thalamic nuclei at P0 and P10.
a, ‘Leave-one-out’ cross-validation analysis confirms the robustness of the support vector machine model at P3. See Methods for details. b, FO versus HO delineation is superior to the S versus V delineation at all levels of stringency. c, Type-specific genes are more differentially expressed between FO and HO nuclei than between S and V nuclei. Fold changes represent ratios of expression between significantly differentially expressed genes for each condition. *P < 0.05; **P < 0.01; Welch’s two-sample t-test.
Extended Data Figure 2 Hierarchical order-based transcriptional logic applies across sensory modalities.
a, Top, schematic representation of auditory pathways. Bottom left, anterograde labelling from vMG and dMG. Note that in contrast to somatosensory and visual modalities, both nuclei project to L1 and avoid L5A. Bottom right, Illustrative microdissection, acute coronal section. Nuclei were identified by retrograde labelling from A1. b, Unbiased clustering delineates FO and HO nuclei. Shaded ellipse represents 85% confidence area around centroid. Circles represent individual samples. c, Expression of the FO- and HO-specific transcripts (‘FO genes’, ‘HO genes’) in distinct sensory nuclei. Error bars in the right panel indicate s.e.m. d, Unbiased classification using vMG- and dMG-specific transcripts as training sets showed a corresponding hierarchical order-based distribution of somatosensory and visual nuclei. e, f, In situ hybridization on coronal sections showing 8 FO (e) and 13 HO-specific transcripts (f) and their corresponding level of expression in microarray data. P4 in situ hybridization of Slc6a4, Gbx2, Calb2 and Cdhr1 are from the Allen Brain Atlas. P7 in situ hybridization of Sorbs1, Dcdc2a, Gria2, Adarb1, Nefm, Plxcn1, Lypd1, Adcyap1, Sstr4, Prox1, Caln1 and Cit are from the St Jude Brain Gene Expression Map (BGEM, hosted at http://www.gensat.org)31. In situ hybridization for Id2, Cdkn1c, Glra3, Nxph1 and Tcf7l2 are not available within these databases and were performed based on their high fold-change in gene expression in FO versus HO. Scale bar, 200 μm.
Extended Data Figure 3 Acquisition of final FO neuron identity is a periphery-dependent process.
a, Expression of VB and LG peripheral input-dependent genes (highlighted with an asterisk in Fig. 2). b, Top, IONS leads to decreased expression of VB-type genes and increased expression of Po-type genes. Bottom, Enucleation (enu) leads to decreased expression of LG-type genes and increased expression of LP-type genes. c, IONS and enucleation affect overlapping sets of genes. Top, genes whose expression is modified by IONS are congruently affected by enucleation. Bottom, genes whose expression is modified by enucleation are congruently affected by IONS. Expression of input-dependent genes in both condition is thus highly correlated (VBIONS: P < 0.0001; LGenu P < 0.0001). r, regression coefficient. d, P0 IONS or enucleation prevents the induction of transcriptional programs normally observed between P0 and P3. Linear model using gene expression in control VB at P0 and P3 or control LG at P0 and P3 as training sets.
Extended Data Figure 4 HO nucleus identity is largely independent of peripheral input.
a, P0 IONS or enucleation does not detectably affect gene expression in the Po and LP. Linear model using gene expression in corresponding control FO and HO nuclei as training sets. b, Expression of peripheral input-dependent genes in the Po and LP. c, Quantification of the data shown in b. L, left; R, right; Ctl, control; n.s., not significant. Top, **P < 0.01 (Fisher’s exact test); bottom, P = n.s. (Welch’s two-sample t-test). d, Input ablation does not affect HO developmental dynamics. P = n.s. (Welch’s two-sample t-test). e, IONS and enucleation affect distinct sets of genes. Top, genes with expression that is modified by IONS are not affected by enucleation. Bottom, genes with expression that is modified by enucleation are not affected by IONS. Expression of input-dependent genes in both condition is thus not correlated (top: PoIONS r = −0.33, P < 0.0001; bottom: LPenu r = −0.11, P = not significant).
Extended Data Figure 5 Probing L5B connectivity in Rbp4-Cre mice.
a, Top, schematic representation of the experimental setting and hypothesis. Centre, bottom, the presynaptic terminals of L5B neurons were revealed by crossing Rbp4-Cre mice, in which Cre recombinase is selectively expressed by L5B neurons19, with floxed mutants expressing a tomato-red-tagged version of the presynaptic protein synaptophysin (Syp)32. In contrast to normal LG, L5B presynaptic terminals are present in LG following enucleation. b, Top, Rbp4-Cre+ L5B neurons express mCherry. Bottom left, Burst-firing of L5B mCherry+ neurons. Bottom right: example of photocurrents recorded in voltage-clamp during continuous blue-light stimulation and action potentials induced by blue light stimulation. c, Optogenetic stimulation of primary visual cortex L5B input elicits postsynaptic responses in LP control and LPenu. Percentage of connectivity of LP versus LPenu: not significant; χ2 statistics on contingency table. EPSC amplitude (pA): LP (n = 14 out of 20), 353.1 ± 94.6; LPenu (n = 22 out of 22), 365.5 ± 57.9; LG (n = 1 out of 23), 8.6; LGenu (n = 10 out of 20), 24.5 ± 13.7. Values are shown as mean ± s.e.m.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-2 and an additional reference. (PDF 118 kb)
Supplementary Table 1
This file contains FO/HO- and Somatosensory/Visual-type transcripts. (XLSX 140 kb)
Supplementary Table 2
This file contains VP/Po, LG/LP, and vMG/dMG-type transcripts. (XLSX 185 kb)
Supplementary Table 3
This file shows input-dependent transcripts in the somatosensory and visual system. (XLSX 168 kb)
Rights and permissions
About this article
Cite this article
Frangeul, L., Pouchelon, G., Telley, L. et al. A cross-modal genetic framework for the development and plasticity of sensory pathways. Nature 538, 96–98 (2016). https://doi.org/10.1038/nature19770
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19770
This article is cited by
-
A Thalamocortical Perspective on Sleep Spindle Alterations in Neurodevelopmental Disorders
Current Sleep Medicine Reports (2024)
-
Modular strategy for development of the hierarchical visual network in mice
Nature (2022)
-
Evolution of central neural circuits: state of the art and perspectives
Nature Reviews Neuroscience (2022)
-
Precise coupling of the thalamic head-direction system to hippocampal ripples
Nature Communications (2020)
-
Anatomically and functionally distinct thalamocortical inputs to primary and secondary mouse whisker somatosensory cortices
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.