Abstract
In Gram-negative bacteria, outer membrane transporters import nutrients by coupling to an inner membrane protein complex called the Ton complex. The Ton complex consists of TonB, ExbB, and ExbD, and uses the proton motive force at the inner membrane to transduce energy to the outer membrane via TonB. Here, we structurally characterize the Ton complex from Escherichia coli using X-ray crystallography, electron microscopy, double electron–electron resonance (DEER) spectroscopy, and crosslinking. Our results reveal a stoichiometry consisting of a pentamer of ExbB, a dimer of ExbD, and at least one TonB. Electrophysiology studies show that the Ton subcomplex forms pH-sensitive cation-selective channels and provide insight into the mechanism by which it may harness the proton motive force to produce energy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
05 October 2016
A minor change was made to the reported labelling at residue 113 distance, and to the DEER Spectroscopy section in the Methods.
References
Krewulak, K. D. & Vogel, H. J. TonB or not TonB: is that the question? Biochem. Cell Biol. 89, 87–97 (2011)
Noinaj, N., Guillier, M., Barnard, T. J. & Buchanan, S. K. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 (2010)
Bassford, P. J., Jr, Bradbeer, C., Kadner, R. J. & Schnaitman, C. A. Transport of vitamin B12 in tonB mutants of Escherichia coli. J. Bacteriol. 128, 242–247 (1976)
Lohmiller, S., Hantke, K., Patzer, S. I. & Braun, V. TonB-dependent maltose transport by Caulobacter crescentus. Microbiology 154, 1748–1754 (2008)
Schauer, K., Rodionov, D. A. & de Reuse, H. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338 (2008)
Cascales, E. et al. Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 (2007)
Postle, K. & Kadner, R. J. Touch and go: tying TonB to transport. Mol. Microbiol. 49, 869–882 (2003)
Clément, E., Mesini, P. J., Pattus, F. & Schalk, I. J. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43, 7954–7965 (2004)
Cadieux, N., Barekzi, N. & Bradbeer, C. Observations on the calcium dependence and reversibility of cobalamin transport across the outer membrane of Escherichia coli. J. Biol. Chem. 282, 34921–34928 (2007)
Schramm, E., Mende, J., Braun, V. & Kamp, R. M. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J. Bacteriol. 169, 3350–3357 (1987)
Brewer, S. et al. Structure and function of X-Pro dipeptide repeats in the TonB proteins of Salmonella typhimurium and Escherichia coli. J. Mol. Biol. 216, 883–895 (1990)
Chang, C., Mooser, A., Plückthun, A. & Wlodawer, A. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276, 27535–27540 (2001)
Witty, M. et al. Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein. EMBO J. 21, 4207–4218 (2002)
Ködding, J. et al. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J. Biol. Chem. 280, 3022–3028 (2005)
Garcia-Herrero, A., Peacock, R. S., Howard, S. P. & Vogel, H. J. The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins. Mol. Microbiol. 66, 872–889 (2007)
Higgs, P. I., Larsen, R. A. & Postle, K. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44, 271–281 (2002)
Sverzhinsky, A. et al. Membrane protein complex ExbB4-ExbD1-TonB1 from Escherichia coli demonstrates conformational plasticity. J. Bacteriol. 197, 1873–1885 (2015)
Sverzhinsky, A. et al. Coordinated rearrangements between cytoplasmic and periplasmic domains of the membrane protein complex ExbB-ExbD of Escherichia coli. Structure 22, 791–797 (2014)
Pramanik, A. et al. Oligomeric structure of ExbB and ExbB-ExbD isolated from Escherichia coli as revealed by LILBID mass spectrometry. Biochemistry 50, 8950–8956 (2011)
Jordan, L. D. et al. Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc. Natl Acad. Sci. USA 110, 11553–11558 (2013)
Braun, V. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 171, 6387–6390 (1989)
Cascales, E., Lloubès, R. & Sturgis, J. N. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42, 795–807 (2001)
Lloubès, R. et al. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152, 523–529 (2001)
Yeh, Y. C., Comolli, L. R., Downing, K. H., Shapiro, L. & McAdams, H. H. The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J. Bacteriol. 192, 4847–4858 (2010)
Gray, A. N. et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4, 07118 (2015)
Germon, P., Ray, M. C., Vianney, A. & Lazzaroni, J. C. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183, 4110–4114 (2001)
Larsen, R. A., Thomas, M. G. & Postle, K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31, 1809–1824 (1999)
Zhai, Y. F., Heijne, W. & Saier, M. H., Jr. Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta 1614, 201–210 (2003)
Kojima, S. & Blair, D. F. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40, 13041–13050 (2001)
Saier, M. H., Jr. Tracing pathways of transport protein evolution. Mol. Microbiol. 48, 1145–1156 (2003)
Ollis, A. A., Manning, M., Held, K. G. & Postle, K. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73, 466–481 (2009)
Parsons, L. M., Grishaev, A. & Bax, A. The periplasmic domain of TolR from Haemophilus influenzae forms a dimer with a large hydrophobic groove: NMR solution structure and comparison to SAXS data. Biochemistry 47, 3131–3142 (2008)
Larsen, R. A. & Postle, K. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276, 8111–8117 (2001)
Labarca, P. & Latorre, R. Insertion of ion channels into planar lipid bilayers by vesicle fusion. Methods Enzymol. 207, 447–463 (1992)
Ollis, A. A., Kumar, A. & Postle, K. The ExbD periplasmic domain contains distinct functional regions for two stages in TonB energization. J. Bacteriol. 194, 3069–3077 (2012)
Ollis, A. A. & Postle, K. ExbD mutants define initial stages in TonB energization. J. Mol. Biol. 415, 237–247 (2012)
Braun, V. et al. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178, 2836–2845 (1996)
Braun, V. & Herrmann, C. Point mutations in transmembrane helices 2 and 3 of ExbB and TolQ affect their activities in Escherichia coli K-12. J. Bacteriol. 186, 4402–4406 (2004)
Sehnal, D. et al. MOLE 2.0: advanced approach for analysis of biomacromolecular channels. J. Cheminform. 5, 39 (2013)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996)
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012)
Whitmore, L. & Wallace, B. A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 (2008)
Minor, Z. O. W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010)
Schneider, T. P. T. R. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Pettersen, E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Schenk, A. D., Philippsen, A., Engel, A. & Walz, T. A pipeline for comprehensive and automated processing of electron diffraction data in IPLT. J. Struct. Biol. 182, 173–185 (2013)
Polyhach, Y. et al. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 14, 10762–10773 (2012)
Jeschke, G. et al. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498 (2006)
von Hagens, T., Polyhach, Y., Sajid, M., Godt, A. & Jeschke, G. Suppression of ghost distances in multiple-spin double electron-electron resonance. Phys. Chem. Chem. Phys. 15, 5854–5866 (2013)
Polyhach, Y., Bordignon, E. & Jeschke, G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 13, 2356–2366 (2011)
Mueller, P., Rudin, D. O., Tien, H. T. & Wescott, W. C. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature 194, 979–980 (1962)
Hille, B. Ion Channels of Excitable Membranes 3rd edn (Sinauer, 2001)
Acknowledgements
We thank the staff at the SER-CAT and GM/CA-CAT beamlines at the Advanced Photon Source (APS), Argonne National Laboratory (use of the APS is supported by the US DOE, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38, and by the US DOE, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357); the staff at beamlines 5.0.1 and 8.2.1, Advance Light Source at Lawrence Berkeley National Laboratory for their assistance during crystal screening (the Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US DOE under Contract No. DE-AC02-05CH11231); G. Jeschke (ETH Zurich) for providing the Q-band resonator; T. Assafa for the reproducibility measurements; the Deutsche Forschungsgemeinschaft for funding the AWG E580 Q-band spectrometer (INST 130/972-1 FUGG); Y. Li at the NINDS/NIH Protein/Peptide Sequencing Facility for performing mass spectrometry analysis; and members of the Lloubes team, E. Cascales, J. Sturgis and J. P. Duneau for discussions. N.N. is supported by the Department of Biological Sciences, Purdue University and by the National Institute of Allergy and Infectious Diseases (1K22AI113078-01). E.B. is supported by the Cluster of Excellence RESOLV (EXC 1069) funded by the Deutsche Forschungsgemeinschaft. W.A.C. is supported by the NIH (NIH GM 038323) and the Henry Koffler Professorship at Purdue University. H.C. and R.L. are funded by the Centre National de la Recherche Scientifique, the Aix-Marseille Université and grants from the Agence National de la Recherche (BACMOLMOT [ANR-14-CE09-0023]) and from Projets internationaux de coopération scientifique (PICS05853). H.C., T.J.B., I.B. and S.K.B. are supported by the Intramural Research Program of the NIH, NIDDK.
Author information
Authors and Affiliations
Contributions
M.S. prepared the ExbB wild-type construct. H.C., N.N., T.J.B., and R.L. prepared ExbD constructs and mutants of ExbB and ExbD. T.J.B. prepared the TonB constructs. H.C. performed all expression and purification for assays, electron microscopy, DEER spectroscopy, electrophysiology, and crystallization. H.C. and N.N. performed crystallization and H.C., N.N., and I.B. did screening and data collection. N.N. solved the initial crystal structure using crystals grown by H.C. H.C. performed the electron microscopy experiments. E.B. performed the DEER spectroscopy experiments. S.D.Z. and W.A.C. performed the electrophysiology experiments. All authors analysed and discussed the data. R.L., W.A.C., and S.K.B. conceived the original projects. H.C., N.N., S.D.Z., W.A.C., R.L. and S.K.B. contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks H. Mchaourab, T. Walz and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
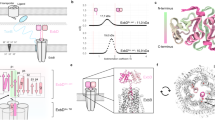
Extended Data Figure 1 Structure determination for the Ton subcomplex (ExbB–ExbDΔperi) using Se-SAD at 5.2 Å resolution.
a, The initial structure of the Ton subcomplex was solved by Se-SAD using anisotropic data extending to 5.2 Å resolution. The data from two crystals were processed with Xia2 and the initial sites found using HKL2MAP v0.3, which found a single solution every ~10,000 tries; resolution limits were also important for finding a solution. b, The sites were then input into AUTOSOL/PHENIX for site refinement and density modification, producing density maps (blue mesh) which clearly showed five-fold symmetry and allowed an initial model of a monomer to be built, consisting almost entirely of α-helices. This model was then used as a search model for molecular replacement to solve the 2.6 Å native structure (data obtained from a single crystal). c, Anomalous different map (orange mesh) showing density for the selenium sites in the 5.2 Å Se-incorporated structure.
Extended Data Figure 2 Representative electron density for the native Ton subcomplex (ExbB–ExbDΔperi) solved to 2.6 Å resolution.
a, Representative electron density map (2Fo−Fc contoured at 1.0σ, grey mesh; 2Fo−Fc omit map (omitting residues 113–124) contoured at 1.0σ, magenta mesh) along residues 113–124 within helix α5. b, Cutaway view of the transmembrane pore of ExbB (grey ribbon) from the native structure at pH 7.0 showing ring-like difference density (green isosurface, Fo−Fc map contoured at 2.5σ) along the conserved residues T148 and T181 (grey and red spheres). c, d, Tilted view (c) and an orthogonal view (d) (relative to a) of the ring-like density. Structures were determined using data obtained from a single crystal in each case.
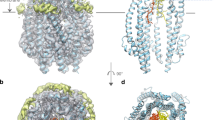
Extended Data Figure 3 Helical shifts and overall flexibility in the ExbB pentamer.
a, Two pentamers were observed per asymmetric unit within the crystal structure. Shown here is pentamer 1 (green) aligned with pentamer 2 (magenta), illustrating slight shifts in a number of the helices (cylinders) between the two pentamers, with the largest shifts indicated by black arrows. The loops connecting α6 and α7 also show variability between monomers and pentamers. b, The TonB subcomplex (ExbB–ExbDΔperi) showing a B-factor putty representation with values ranging from the most ordered in blue to the most disordered in red.
Extended Data Figure 4 Electron density for the transmembrane helix of ExbD.
a, Omit map (2Fo−Fc, contoured at 1.0σ) along the transmembrane pore of ExbB. The density corresponding to the ExbB pentamer is shown in blue mesh, while the density corresponding to the transmembrane helix of ExbD is shown in green mesh. b, Stereoimage showing the density (2Fo−Fc, contoured at 0.8σ, grey mesh; 2Fo−Fc omit map (omitting the transmembrane helix of ExbD), contoured at 0.8σ, green mesh) for the transmembrane helix of ExbD after building and refinement.
Extended Data Figure 5 Comparison of observed density for crystal structures of ExbB–ExbDΔperi solved at pH 7.0 versus pH 4.5.
The presence of electron density for the transmembrane helix of ExbD (magenta ribbon) was dependent on the pH at which the crystals were grown. At pH 7.0, we observed little density (orange mesh) inside the transmembrane pore of the ExbB (grey ribbon) pentamer (see also Extended Data Fig. 3). However, for the structures solved at pH 4.5, we observed clear density (blue mesh) for the transmembrane helix of ExbD, albeit to varying degrees. Density maps (2Fo−Fc) are contoured at 1.0σ.
Extended Data Figure 6 Packing similarities of the 2D and 3D crystals used for electron microscopy and X-ray crystallography.
a, Averaged projection map from the electron microscopy analysis on 2D crystals. Five images were analysed, and a representative averaged projection map was calculated from 900 sub-images. The averaged map shows two different populations of the pentamer that are similar in size but differ in level intensity owing to opposite orientations of the complex within the crystal; a similar packing arrangement was also observed in our crystal structures. ExbD was not detected in our electron microscopy studies, probably owing to disorder of the globular domain, which is anchored to the membrane by a long unstructured linker15. b, Packing of the complex in the X-ray crystal structure from 3D crystals. The right side indicates an orthogonal view highlighting a single row of molecules from the lattice (black dashed box). c, Fitting the row of molecules from the 3D lattice (X-ray) from b onto the averaged projection map from the 2D crystals (electron microscopy) to highlight the consistency observed in packing.
Extended Data Figure 7 DEER traces and analysis.
Ton subcomplex (ExbBC25–ExbD, ExbBC25S–ExbDN78C, and ExbBC25S–ExbDE113C) in 0.08% C10E5 (a) and in 0.03% DDM (b), and the fully assembled Ton complex (TonBC18A–ExbBC25–ExbD and TonBC18A–ExbBC25S–ExbDN78C) in 0.05% DDM (c). Upper panels, experimental Q-band DEER primary data V(t)/V(0) (coloured lines, cyan ExbD113MTSL; violet ExbD78MTSL; red and orange, ExbB25MTSL) and simulated background functions (dotted line). Middle panels, DEER traces after background correction (coloured lines) and fit with DeerAnalysis2015 (dotted lines) with Tikhonov regularization parameters from 10 to 100 adjusted via L-curve analysis and data validation. Lower panels, obtained distance distributions. For the pentameric ExbB sample (50% labelling efficiency), a modulation depth >0.45 was obtained, indicating the presence of a multi-spin system. For the sample solubilized in DDM, longer DEER traces were obtained (4 μs) to better characterize the long distance peak of 5–6 nm in ExbB25MTSL. Additionally, for all panels, another DEER trace was measured after decreasing the microwave power of the 12-ns pump pulse to 25% (orange line) to suppress ghost peaks arising from the presence of more than two spins in the system. The resulting distance distribution (orange) was found to be very similar to that obtained with 100% microwave power (red), showing that no ghost peak artefacts were present. The lower modulation depth observed for the ExbD samples labelled at position 113 with respect to those labelled at position 78 (both labelling efficiency >80%) may be due to the presence of distances <1.5 nm (predicted by the simulations), which are outside of the sensitivity range of the technique, or to destabilization of the ExbD dimer induced by the label. The bottom of c shows a comparison of the Ton subcomplex in DDM (dashed lines from b) to the fully assembled Ton complex (solid lines). All panels show data from single experiments.
Extended Data Figure 8 Densitometry of the purified fully assembled Ton complex.
a, SDS–PAGE gel of the Ton complex (+TonB) and the Ton subcomplex (−TonB) at increasing concentrations. b, Bar graph showing the comparison of the ExbB–ExbD ratio within the Ton complex (+TonB) and the Ton subcomplex (−TonB) indicating that association of TonB with the Ton subcomplex does not change the stoichiometric ratio of the components. While we see a slight difference in the ExbB–ExbD ratio values in the presence or absence of TonB, the observed difference is too small to suggest an altered stoichiometry between ExbB and ExbD. Three representative lanes for each sample are shown in a; however, five lanes were used for all calculations. Densitometry analysis was performed with ImageJ and mean values and standard errors calculated using Microsoft Excel. For purifications of the Ton complex (+TonB), five purification experiments were performed and one representative is shown. For purifications of the Ton subcomplex (−TonB), ~50 purifications were performed and one representative is shown.
Extended Data Figure 9 Circular dichroism analysis of secondary structure and thermal stability of the Ton subcomplex.
Far-UV circular dichroism spectrum (185–260 nm) of the Ton subcomplex (ExbB–ExbD) with the calculated percentage of secondary structure shown. Contents of regular and distorted α-helical structures, 47 and 21%, respectively, were combined during the calculation of secondary structure contributions. Inset, comparison of the thermal stability of the Ton subcomplex (blue) versus ExbB alone (red) measured through the temperature dependence of the circular dichroism signal amplitude at 222 nm. Both panels show data from a single experiment.
Extended Data Figure 10 Sequence conservation of ExbB orthologues mapped onto the crystal structure.
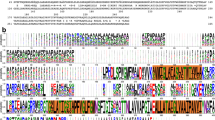
a, Clustal W alignment of ExbB sequences from: E.coli K12 (P0ABU7), Neisseria meningitidis (P64100), Neisseria gonorrhoeae (Q5F711), Haemophilus ducreyi (O51808), Vibrio harveyi (D0XEN5), Yersinia pestis (D1TTA4), Methanothermobacter thermautotrophicus (O27101), Pseudomonas aeruginosa (G3XCW0), ExbB1 of Vibrio cholerae (O52043) and ExbB2 of Vibrio cholerae (AAC69454). b, Conservation mapped onto the ExbB structure with Chimera. The most conserved residues are in blue and found in α6 (TM2) and α7 (TM3) of the ExbB structure. An extensive alignment that also includes sequences from the Tol and Mot systems shows similar results22. c, Cutaway molecular surface of ExbB pentamer with the most conserved residues mapped onto the surface.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-2. Table 1 contains a summary of data collection and refinement statistics for the Ton subcomplex crystal structures; Table 2 shows how zero-current potential (Vrev) was determined from volt-ampere characteristics measured in asymmetric salt conditions. Relative cation/anion permeability was calculated using the Goldman-Hodgkin-Katz equation. (XLSX 12 kb)
Overall structure of the Ton subcomplex
This video gives a structural insight into the role of the Ton complex in energy transduction. (MOV 29929 kb)
Rights and permissions
About this article
Cite this article
Celia, H., Noinaj, N., Zakharov, S. et al. Structural insight into the role of the Ton complex in energy transduction. Nature 538, 60–65 (2016). https://doi.org/10.1038/nature19757
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19757
This article is cited by
-
Bacteroides ovatus accelerates metformin-induced vitamin B12 deficiency in type 2 diabetes patients by accumulating cobalamin
npj Biofilms and Microbiomes (2023)
-
The TonB system in Aeromonas hydrophila NJ-35 is essential for MacA2B2 efflux pump-mediated macrolide resistance
Veterinary Research (2021)
-
Functional roles of multiple Ton complex genes in a Sphingobium degrader of lignin-derived aromatic compounds
Scientific Reports (2021)
-
The lipoprotein Pal stabilises the bacterial outer membrane during constriction by a mobilisation-and-capture mechanism
Nature Communications (2020)
-
The importance of the membrane for biophysical measurements
Nature Chemical Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.