Abstract
Little is known about the ability of Drosophila circadian neurons to promote sleep. Here we show, using optogenetic manipulation and video recording, that a subset of dorsal clock neurons (DN1s) are potent sleep-promoting cells that release glutamate to directly inhibit key pacemaker neurons. The pacemakers promote morning arousal by activating these DN1s, implying that a late-day feedback circuit drives midday siesta and night-time sleep. To investigate more plastic aspects of the sleep program, we used a calcium assay to monitor and compare the real-time activity of DN1 neurons in freely behaving males and females. Our results revealed that DN1 neurons were more active in males than in females, consistent with the finding that male flies sleep more during the day. DN1 activity is also enhanced by elevated temperature, consistent with the ability of higher temperatures to increase sleep. These new approaches indicate that DN1s have a major effect on the fly sleep–wake profile and integrate environmental information with the circadian molecular program.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Reppert, S. M. & Weaver, D. R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002)
Moore, R. Y. Organization and function of a central nervous system circadian oscillator: the suprachiasmatic hypothalamic nucleus. Fed. Proc. 42, 2783–2789 (1983)
Rieger, D., Shafer, O. T., Tomioka, K. & Helfrich-Förster, C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster . J. Neurosci. 26, 2531–2543 (2006)
Shang, Y., Griffith, L. C. & Rosbash, M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl Acad. Sci. USA 105, 19587–19594 (2008)
Cavanaugh, D. J. et al. Identification of a circadian output circuit for rest:activity rhythms in Drosophila . Cell 157, 689–701 (2014)
Agosto, J. et al. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila . Nat. Neurosci. 11, 354–359 (2008)
Tataroglu, O. & Emery, P. Studying circadian rhythms in Drosophila melanogaster . Methods 68, 140–150 (2014)
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C. & Taghert, P. H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila . Cell 99, 791–802 (1999)
Grima, B., Chélot, E., Xia, R. & Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873 (2004)
Guo, F., Cerullo, I., Chen, X. & Rosbash, M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila . eLife 3, e02780 (2014)
Yao, Z. & Shafer, O. T. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520 (2014)
Stoleru, D., Peng, Y., Agosto, J. & Rosbash, M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila . Nature 431, 862–868 (2004)
Kunst, M. et al. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila . Curr. Biol. 24, 2652–2664 (2014)
Gilestro, G. F. & Cirelli, C. pySolo: a complete suite for sleep analysis in Drosophila . Bioinformatics 25, 1466–1467 (2009)
Donelson, N. C. et al. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One 7, e37250 (2012)
Garbe, D. S. et al. Context-specific comparison of sleep acquisition systems in Drosophila . Biol. Open 4, 1558–1568 (2015)
Pfeiffer, B. D. et al. Tools for neuroanatomy and neurogenetics in Drosophila . Proc. Natl Acad. Sci. USA 105, 9715–9720 (2008)
Zhang, L. et al. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila . Curr. Biol. 20, 591–599 (2010)
Zhang, Y., Liu, Y., Bilodeau-Wentworth, D., Hardin, P. E. & Emery, P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr. Biol. 20, 600–605 (2010)
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2, 991–1001 (2012)
Flourakis, M. et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848 (2015)
Seluzicki, A. et al. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 12, e1001810 (2014)
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014)
Aso, Y. et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila . eLife 3, e04580 (2014)
Inagaki, H. K. et al. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods 11, 325–332 (2014)
Gradinaru, V. et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010)
Rieger, D. et al. The fruit fly Drosophila melanogaster favors dim light and times its activity peaks to early dawn and late dusk. J. Biol. Rhythms 22, 387–399 (2007)
Feinberg, E. H. et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008)
Yao, Z., Macara, A. M., Lelito, K. R., Minosyan, T. Y. & Shafer, O. T. Analysis of functional neuronal connectivity in the Drosophila brain. J. Neurophysiol. 108, 684–696 (2012)
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013)
Hamasaka, Y. et al. Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J. Comp. Neurol. 505, 32–45 (2007)
Diao, F. et al. Plug-and-play genetic access to Drosophila cell types using exchangeable exon cassettes. Cell Reports 10, 1410–1421 (2015)
Liu, W. W. & Wilson, R. I. Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc. Natl Acad. Sci. USA 110, 10294–10299 (2013)
Abruzzi, K., Chen, X., Nagoshi, E., Zadina, A. & Rosbash, M. RNA-seq profiling of small numbers of Drosophila neurons. Methods Enzymol. 551, 369–386 (2015)
Collins, B. et al. Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLoS Biol. 12, e1001959 (2014)
Collins, B., Kane, E. A., Reeves, D. C., Akabas, M. H. & Blau, J. Balance of activity between LNvs and glutamatergic dorsal clock neurons promotes robust circadian rhythms in Drosophila . Neuron 74, 706–718 (2012)
Helfrich-Förster, C. Differential control of morning and evening components in the activity rhythm of Drosophila melanogaster—sex-specific differences suggest a different quality of activity. J. Biol. Rhythms 15, 135–154 (2000)
Low, K. H., Lim, C., Ko, H. W. & Edery, I. Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron 60, 1054–1067 (2008)
Cao, W. & Edery, I. A novel pathway for sensory-mediated arousal involves splicing of an intron in the period clock gene. Sleep 38, 41–51 (2015)
Majercak, J., Sidote, D., Hardin, P. E. & Edery, I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230 (1999)
Masuyama, K., Zhang, Y., Rao, Y. & Wang, J. W. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J. Neurogenet. 26, 89–102 (2012)
Stanewsky, R. et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila . Cell 95, 681–692 (1998)
Hardin, P. E., Hall, J. C. & Rosbash, M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc. Natl Acad. Sci. USA 89, 11711–11715 (1992)
Benito, J., Zheng, H., Ng, F. S. & Hardin, P. E. Transcriptional feedback loop regulation, function, and ontogeny in Drosophila . Cold Spring Harb. Symp. Quant. Biol. 72, 437–444 (2007)
Picot, M., Klarsfeld, A., Chélot, E., Malpel, S. & Rouyer, F. A role for blind DN2 clock neurons in temperature entrainment of the Drosophila larval brain. J. Neurosci. 29, 8312–8320 (2009)
Prober, D. A., Rihel, J., Onah, A. A., Sung, R. J. & Schier, A. F. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 26, 13400–13410 (2006)
Liang, X., Holy, T. E. & Taghert, P. H. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo . Science 351, 976–981 (2016)
Gao, X. J. et al. A transcriptional reporter of intracellular Ca2+ in Drosophila . Nat. Neurosci. 18, 917–925 (2015)
Stanewsky, R. Analysis of rhythmic gene expression in adult Drosophila using the firefly luciferase reporter gene. Methods Mol. Biol. 362, 131–142 (2007)
Tang, C. H., Hinteregger, E., Shang, Y. & Rosbash, M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron 66, 378–385 (2010)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Haynes, P. R., Christmann, B. L. & Griffith, L. C. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster . eLife 4, e03868 (2015)
Acknowledgements
We thank M. Diaz, N. Nguyen, R. Spann and K. Kerr for generous help, and X. Gao and L. Luo for sending us LexAop-LUC flies, and O. Shafer and A. Sehgal for helpful discussion and comments on early versions of this manuscript. This work was supported in part by NIH R01 MH067284 (LCG).
Author information
Authors and Affiliations
Contributions
F.G. and M.R. conceived and designed the experiments. F.G. performed behavioural experiments. F.G. and J.Y. performed immunocytochemistry and imaging experiments. F.G. and H.J.J. setup the recording system. K.C.A. and W.L. performed and quantified the mRNA profiling data. F.G. and J.Y. analysed data. F.G., L.G. and M.R. prepared the figures and wrote the paper, with feedback from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks A. Sehgal and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Schematics of video recording and optogenetic strategies.
a, Flies expressing CsChrimson were placed in 96-well plates and video recorded with a camera without an infrared filter (left). An 850 nm infrared back light provides illumination for recording in both light and dark periods. A set of 627 nm LEDs was carefully positioned and combined with a diffuser to ensure uniform irradiation for stimulation. The voltage and pulse frequency were controlled by an Arduino UNO board as described in Methods. Representative data from a video recording of male and female activity (right) in light:dark are shown. b, Sleep data of two control genotypes in 96-well plate mode for 6 days. Results with error bars are mean ± s.e.m. n = 15–16 for each group.
Extended Data Figure 2 The R18H11 driver labels a subgroup of CLK4.1M-defined DN1s.
Confocal stack of images showing antibody staining for GFP (left) and RFP (middle) and the overlay (right) in the dorsal brain of R18H11-LexA/LexAop-mCD8::GFP;Clk4.1M-GAL4/UAS-rCD2::RFP flies. Scale bar, 20 μm.
Extended Data Figure 3 Blocking neurotransmitter output of DN1s affects sleep parameters but not DD rhythmicity.
a–f, Total sleep, maximum sleep bout duration and mean sleep bout duration of control groups (R18H11-GAL4/+, UAS-TNT/+) and the experimental group (R18H11-GAL4/UAS-TNT) have significant differences. Results with error bars are mean ± s.e.m. n = 32 for each group. One-way ANOVA was performed to detect significant genotype effects for total sleep (P < 0.0001), daytime sleep (P < 0.0001), night-time sleep (P < 0.0001) (a, b), maximum bout duration (P = 0.00288), maximum daytime bout duration (P < 0.0001), maximum night-time bout duration (P = 0.000388) (d), mean bout duration (P < 0.0001), mean daytime bout duration (P < 0.0001), mean night-time bout duration (P < 0.0001) (e). Asterisks denote significant differences from parental controls in Tukey’s post-hoc test (P < 0.01). g, Neurotransmitter release from DN1s is not required for DD rhythmicity. Locomotor behaviour of control (Clk4.1M-GAL4/UAS-Tet) and experimental (Clk4.1M-GAL4/UAS-TNT) male flies was monitored for 6 days in DD. Both control (left) and experimental (right) flies maintained strong rhythmicity. Note that the experimental group showed much less daytime sleep and higher activity level (dashed red arrow-right panel) than the control group (solid red arrow-left panel). n = 32 for each group.
Extended Data Figure 4 Co-expression of CRY-GAL80 or TNT blocks the sleep-promoting effect of DN1 activation.
a, R18H11-GAL4/UAS-CsChrimson (left) and R18H11-GAL4, CRY-GAL80/UAS-CsChrimson (right) brains were dissected and stained with anti-GFP (green). Scale bar, 100 μm. b, Comparison of total sleep in the baseline day (blue) to total sleep during a 24 h LED stimulation day (red) for each genotype. n = 32 for R18H11-GAL4/UAS-CsChrimson and n = 24 for the other groups. Results with error bars are mean ± s.e.m. **P < 0.001 by post-hoc Bonferonni multiple comparisons. Two-way ANOVA was performed to detect a significant LED stimulation effect (P = 0.00227674), a genotype effect (P < 0.0001) and interaction (P < 0.0001).
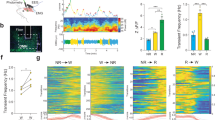
Extended Data Figure 5 Arousal threshold is affected by manipulation of DN1 activity.
a, Mechanical stimulation setup for measuring arousal threshold. The set-up is illustrated on the left. A 96-well plate is loaded onto the device and a small push–pull solenoid is positioned on the side of the plate. The solenoid can be programed to tap the plate at different frequencies and times. A web camera monitors fly movement in the wells, and the video is analysed with Fiji ImageJ software to track flies. An example image is shown on the right. b, Activation of DN1s increases arousal threshold. The left panel shows representative trajectories (5 min traces) of experimental and control flies after a strong (10 tap) stimulus at ZT6 when the LED is on. The right panel shows the percentages of flies for the indicated genotypes that transitioned from immobility to an active state in response to the stimulus. The pink background indicates LED stimulation. n = 24 for R18H11-GAL4/UAS-CsChrimson and R18H11-GAL4/UAS-TNT groups, n = 16 for parental control groups. Results with error bars are mean ± s.e.m. One-way ANOVA was performed to detect significant genotype effects for arousal levels of R18H11>CsChrimson LED group (P < 0.0001) and R18H11>TNT group (P < 0.0001). Asterisks denote significant differences from parental controls in Tukey’s post-hoc test (P < 0.01).
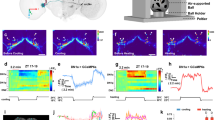
Extended Data Figure 6 eNPHR3.0 blocks DN1 neuronal activity and decreases the siesta.
a, The luminescence traces from the CaLexA–LUC sensor reflect neuronal activity after LED-induced eNPHR3.0 inhibition from ZT 2.5 to ZT 9.5. The mean LUC activity level (arbitrary units) from control and experimental groups is quantified on the right. Results with shading are mean ± s.e.m. Box boundaries represent the first and third quartiles, whiskers are 1.5 interquartile range. n = 24 for each group. b, Sleep from a baseline day and from a LED stimulation day of R18H11>UAS-eNPHR3.0, UAS-eNPHR3.0/+ and R18H11/+ flies. Pink background represents LED stimulation. n = 16 for each group. Results with error bars are mean ± s.e.m.
Extended Data Figure 7 The dendritic region of E cells overlaps with the presynaptic region of DN1s.
Clk4.1M-GAL4>UAS-synaptotagmin-GFP brains were dissected and stained with anti-GFP antibodies to identify the DN1 presynaptic regions (green; left panel). To identify the dendritic regions of E cells and M cells, DvPdf-GAL4>UAS-Denmark brains were dissected and stained with anti-DsRed antibodies (red; middle panel). These patterns were aligned and overlaid (merged panel on the right).
Extended Data Figure 8 Glutamate reduces calcium levels in PDF neurons and hyperpolarizes their membrane potential.
a, Quantification of peak GCaMP6f changes shown in Fig. 2c. Average maximum changes for LNvs and dorsal lateral neurons are shown. n = 8 for control group, 7 for PDF cells and 11 for dorsal lateral neurons. **P < 0.001 by unpaired two-tailed Student’s t-test and results with error bars are mean ± s.e.m. b, Normalized calcium traces in different circadian neuron subgroups imaged concurrently in the same representative brain. c, d, 5 mM glutamate was applied to exposed dissected fly brains (Pdf-GAL4>GCaMP6f, (c) and Pdf-GAL4>Arclight (d)) and induced a calcium decrease and hyperpolarized these core pacemakers (ΔF/F represents the evoked fluorescence change from baseline). The red solid line indicates time of glutamate application. The dashed line indicates time of vehicle application. Panels show representative data of 6 brains.
Extended Data Figure 9 Decreasing DN1 VGLUT levels blocks the E peak reduction caused by DN1 firing.
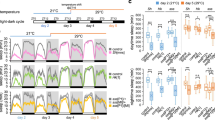
Reducing VGLUT activity within the DN1s decreases the DN1-activation effect (left). The locomotor activity patterns of UAS-dTrpA1/+; CLK4.1M-GAL4/+ female flies (upper panel) and UAS-dTrpA1/+; CLK4.1M-GAL4/UAS-VGlut RNAi female flies (lower panel) at 21 °C and 27 °C are shown. The colour of the bars indicates either daytime (white) or night-time (black). Evening activity index was calculated as described in the Methods for the indicated genotypes (right). White and black bars represent data from low and high temperature respectively. **P < 0.001 by unpaired two-tailed Student’s t-test. n = 24 for each group. Results with error bars are mean ± s.e.m.
Extended Data Figure 10 Reducing mGluRA expression in pacemaker neurons reduces the inhibitory effect of glutamate as well as the siesta.
a, The peak decrease of GCaMP6f in circadian cells after applying glutamate to control and mGluRA knockdown flies. The genotypes are shown above the bars. n = 6 UAS-Dcr2; Clk856-GAL4, UAS-GCaMP6f and n = 8–9 UAS-Dcr2; Clk856-GAL4, UAS-GCaMP6f; UAS-mGluRA RNAi groups. *P < 0.05 by unpaired t-test. b, Comparison of total sleep, daytime siesta and night-time sleep in different genotypes. n = 32 for each group. *P < 0.05 by one-way ANOVA with Tukey’s post-hoc test. Results with error bars are mean ± s.e.m (a, b). c, A temporally constrained negative feedback core pacemaker–DN1 circuit regulates the fly activity–sleep pattern. Early in the day, M pacemaker neurons activate the DN1s via the PDF neuropeptide, and DN1s release DH31 to enhance morning arousal. Later in the day, glutamate release from DN1s inhibits M cells and E cells, promotes the siesta, decreases the evening activity peak and initiates night-time sleep. A cycling mRNA that encodes inhibitory glutamate receptors in pacemaker cells may help direct this inhibition to the late day. This feedback circadian circuit shapes the bimodal locomotor activity peak and sleep–wake cycles under normal conditions. The higher daily neuronal activity in male DN1s compared to female DN1s promotes the sexually dimorphic activity/sleep pattern. DN1s also integrate environmental information such as temperature to promote sleep plasticity.
Supplementary information
Live calcium imaging of circadian neuron responses to DN1 activation
The calcium responses of clock neurons expressing GCaMP6f to ATP/P2X2-activation of DN1s. The genotype of flies used in the experiment is Clk856-GAL4/UAS-GCaMP6f; Clk4.1M-LexA/LexAop-P2X2. This video is accelerated 15X. ATP was added from 30 S (2S in the video). (AVI 20169 kb)
R18H11-GAL4-labeled DN1 arbors co-localize with VGLUT
Confocal stack of antibody staining of GFP and VGLUT (red) in the dorsal brain of R18H11-GAL4>UAS-mCD8::GFP flies. Most R18H11-GAL4-labeled DN1s are glutamatergic as VGLUT is highly enriched in their projections. (AVI 15364 kb)
Rights and permissions
About this article
Cite this article
Guo, F., Yu, J., Jung, H. et al. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536, 292–297 (2016). https://doi.org/10.1038/nature19097
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19097
This article is cited by
-
A four-oscillator model of seasonally adapted morning and evening activities in Drosophila melanogaster
Journal of Comparative Physiology A (2023)
-
Neural Control of Action Selection Among Innate Behaviors
Neuroscience Bulletin (2022)
-
Alterations in the activity and sleep of Drosophila melanogaster under simulated microgravity
npj Microgravity (2021)
-
Metabolic control of daily locomotor activity mediated by tachykinin in Drosophila
Communications Biology (2021)
-
MaxEnt modelling of the potential distribution areas of cultural ecosystem services using social media data and GIS
Environment, Development and Sustainability (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.