Abstract
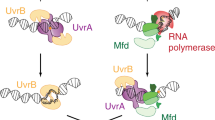
Escherichia coli Mfd translocase enables transcription-coupled repair by displacing RNA polymerase (RNAP) stalled on a DNA lesion and then coordinating assembly of the UvrAB(C) components at the damage site1,2,3,4. Recent studies have shown that after binding to and dislodging stalled RNAP, Mfd remains on the DNA in the form of a stable, slowly translocating complex with evicted RNAP attached5,6. Here we find, using a series of single-molecule assays, that recruitment of UvrA and UvrAB to Mfd–RNAP arrests the translocating complex and causes its dissolution. Correlative single-molecule nanomanipulation and fluorescence measurements show that dissolution of the complex leads to loss of both RNAP and Mfd. Subsequent DNA incision by UvrC is faster than when only UvrAB(C) are available, in part because UvrAB binds 20–200 times more strongly to Mfd–RNAP than to DNA damage. These observations provide a quantitative framework for comparing complementary DNA repair pathways in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Witkin, E. M. Radiation-induced mutations and their repair. Science 152, 1345–1353 (1966)
Mellon, I., Spivak, G. & Hanawalt, P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987)
Selby, C. P. & Sancar, A. Molecular mechanism of transcription-repair coupling. Science 260, 53–58 (1993)
Savery, N. J. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 15, 326–333 (2007)
Howan, K. et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature 490, 431–434 (2012)
Graves, E. T. et al. A dynamic DNA-repair complex observed by correlative single-molecule nanomanipulation and fluorescence. Nature Struct. Mol. Biol . 22, 452–457 (2015)
Deaconescu, A. M. et al. Structural basis for bacterial transcription-coupled DNA repair. Cell 124, 507–520 (2006)
Westblade, L. F. et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 38, 8357–8369 (2010)
Srivastava, D. B. & Darst, S. A. Derepression of bacterial transcription-repair coupling factor is associated with a profound conformational change. J. Mol. Biol. 406, 275–284 (2011)
Park, J.-S., Marr, M. T. & Roberts, J. W. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767 (2002)
Smith, A. J., Szczelkun, M. D. & Savery, N. J. Controlling the motor activity of a transcription-repair coupling factor: autoinhibition and the role of RNA polymerase. Nucleic Acids Res. 35, 1802–1811 (2007)
Haines, N. M., Kim, Y.-I. T., Smith, A. J. & Savery, N. J. Stalled transcription complexes promote DNA repair at a distance. Proc. Natl Acad. Sci. USA 111, 4037–4042 (2014)
Strick, T. R., Allemand, J. F., Bensimon, D., Bensimon, A. & Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996)
Wang, M. D. et al. Force and velocity measured for single molecules of RNA polymerase. Science 282, 902–907 (1998)
Revyakin, A., Liu, C., Ebright, R. H. & Strick, T. R. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006)
Kou, S. C., Cherayil, B. J., Min, W., English, B. P. & Xie, X. S. Single-molecule Michaelis-Menten equations. J. Phys. Chem. B 109, 19068–19081 (2005)
Manelyte, L., Kim, Y.-I. T., Smith, A. J., Smith, R. M. & Savery, N. J. Regulation and rate enhancement during transcription-coupled DNA repair. Mol. Cell 40, 714–724 (2010)
Van Houten, B., Gamper, H., Sancar, A. & Hearst, J. E. DNase I footprint of ABC excinuclease. J. Biol. Chem. 262, 13180–13187 (1987)
Selby, C. P. & Sancar, A. Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat. Res. 236, 203–211 (1990)
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010)
Revyakin, A., Ebright, R. H. & Strick, T. R. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc. Natl Acad. Sci. USA 101, 4776–4780 (2004)
Revyakin, A., Ebright, R. H. & Strick, T. R. Single-molecule DNA nanomanipulation: improved resolution through use of shorter DNA fragments. Nature Methods 2, 127–138 (2005)
Manelyte, L. et al. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair 8, 1300–1310 (2009)
Smith, A. J. & Savery, N. J. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 33, 755–764 (2005)
Revyakin, A., Allemand, J. F., Croquette, V., Ebright, R. H. & Strick, T. R. Single-molecule DNA nanomanipulation: detection of promoter-unwinding events by RNA polymerase. Methods Enzymol. 370, 577–598 (2003)
Duboc, C., Graves, E. T. & Strick, T. R. Simple calibration of TIR field depth using the supercoiling response of DNA. Methods 105, 56–61 (2016)
Verhoeven, E. E. A., Wyman, C., Moolenaar, G. F., Hoeijmakers, J. H. J. & Goosen, N. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J. 20, 601–611 (2001)
van den Broek, B., Noom, M. C. & Wuite, G. J. L. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 33, 2676–2684 (2005)
Acknowledgements
This work was made possible by a China Scholarship Council award to J.F., as well as grants from the French Agence Nationale pour la Recherche (RepOne) and the European Science Foundation (EURYI) to T.R.S., as well as core funding from the CNRS and the University of Paris Diderot. The Strick laboratory is also part of the Programme Equipe Labellisées of the Ligue Contre le Cancer. We thank N. Joly for assistance with protein purification and the Strick laboratory for feedback.
Author information
Authors and Affiliations
Contributions
J.F., M.L.C., N.J.S. and T.R.S. planned experiments; J.F., M.L.C. and T.R.S. prepared reagents; J.F. performed tethered-RNAP, tethered-DNA and NanoCOSM assays; M.L.C. and T.R.S. performed tethered-RNAP assays. J.F., M.L.C. and T.R.S. conducted data analysis, and N.J.S. and T.R.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks S. Deindl, J. Elf and J. Roberts for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Motor properties of the translocating Mfd–RNAP complex as seen in the tethered-RNAP assay.
a, Velocity distribution of translocating Mfd–RNAP in the presence of 2 mM ATP and under a weak opposing load (F = 1 pN). Velocity is measured by fitting a single line segment to an entire translocation time-trace; this is made possible by the fact that velocity is essentially constant over the ~7,000 bp of displacement which constitutes an entire trajectory. The velocity distribution is fitted to a Gaussian, giving a mean velocity of 4.7 ± 1 bp s−1 (SD; n = 99 trajectories). b, Tau plot of inverse velocity of translocating Mfd–RNAP as a function of inverse ATP concentration is well-fitted to a line, indicating Michaelian behaviour with  = 16 ± 0.4 μM (s.e.m.) and
= 16 ± 0.4 μM (s.e.m.) and  = 4.7 ± 0.1 bp s−1 (s.e.m.). Error bars, s.e.m. as determined from at least ten trajectories for each ATP concentration.
= 4.7 ± 0.1 bp s−1 (s.e.m.). Error bars, s.e.m. as determined from at least ten trajectories for each ATP concentration.
Extended Data Figure 2 Resolution of the translocating Mfd–RNAP complex by UvrA or by UvrAB is ATP-dependent as shown by the tethered-RNAP translocation assay.
Down-arrows indicate addition of components as noted and as follows. Beginning with stalled RNAP, we add 100 nM Mfd and 2 mM ATP to form the translocating Mfd–RNAP complex. A wash step using 5 ml of reaction buffer lacking ATP is applied to remove (nearly) all the free ATP in solution, causing the translocating complex to come to a nearly complete halt. Then, (a, b) 50 pM UvrA or (c, d) 50 pM UvrA and 250 nM UvrB is added to the experiment. The complex is stable and release of the magnetic bead is not observed. Further addition of ATP-γ-S (2 mM; see b, d) does not permit bead release. However, final addition of ATP (2 mM) leads to rapid release. Red up-arrows indicate bead release in b, d.
Extended Data Figure 3 Characterization of the long-lived Mfd–RNAP intermediate on 2 kb DNA using the tethered-DNA assay.
a, b, Nanomanipulation time-traces showing pulse-chase measurement of the lifetime of the Mfd–RNAP intermediate for CPD-bearing DNA under conditions of positive (+sc) and negative (−sc) supercoiling, respectively. Down-arrows indicate moments of component addition as noted and as follows. First, we load RNAP onto DNA in standard conditions (25 pM RNAP holoenzyme, 500 nM GreB, and the appropriate nucleotide complement, each present at 200 μM). We then wash out free RNAP with reaction buffer supplemented with 500 nM GreB and the nucleotide complement. We then initiate formation of the intermediate by infusion of the above wash solution supplemented with 100 nM Mfd and 2 mM ATP. For negatively supercoiled DNA, RNAP was loaded under conditions of positive supercoiling before the DNA was returned to negative supercoiling; blue line indicates when DNA supercoiling is changed. Black bar highlights the intermediate state. c, d, Lifetime distributions for the Mfd–RNAP intermediate formed on CPD-bearing DNA under conditions of positive or negative supercoiling, respectively, are well fitted to Gaussian distributions (red lines). For positive supercoiling the mean lifetime of the repair intermediate is 548 ± 37 s (s.e.m., n = 29 events: this distribution is also presented in Fig. 2b), and for negative supercoiling the mean lifetime of the repair intermediate is 556 ± 33 s (s.e.m., n = 21 events). e, f, Lifetime distributions for the Mfd–RNAP intermediate formed on C-less cassette DNA under conditions of positive or negative supercoiling, respectively, are also well fitted to Gaussian distributions. For positive supercoiling the mean lifetime of the repair intermediate is 649 ± 13 s (n = 98 events) and for negative supercoiling the mean lifetime of the repair intermediate is 524 ± 26 s (n = 46 events). Data were fitted over the range delimited by the red lines.
Extended Data Figure 4 Characterization of the Mfd–RNAP intermediate on negatively supercoiled 1 kb DNA in the absence or presence of UvrAB using the tethered-DNA assay.
DNA bears a C-less cassette. a, Nanomanipulation time-trace obtained in continuous-tracking mode in the presence of 25 pM RNAP holoenzyme, 100 nM Mfd, 500 nM GreB, 2 mM ATP, 200 μM UTP and 200 μM GTP. b, As in a but in the added presence of 50 pM UvrA and 250 nM UvrB. c, Lifetime distribution of the Mfd–RNAP intermediate in the absence of UvrA and UvrB has a mean lifetime of 258 ± 17 s (s.e.m., n = 33 events). d, Lifetime distribution of the Mfd–RNAP intermediate in the added presence of 50 pM UvrA and 250 nM UvrB has a mean lifetime of 167 ± 17 s (s.e.m., n = 32 events). Data sets for kinetics were obtained using the pulse-chase methodology.
Extended Data Figure 5 Reduced lifetime of the Mfd–RNAP intermediate in the presence of UvrAB, monitored on 2 kb DNA using the tethered-DNA assay.
a, Nanomanipulation time-trace for positively supercoiled DNA (+sc) bearing a C-less cassette in the presence of 25 pM RNAP holoenzyme, 100 nM Mfd, 50 pM UvrA, 250 nM UvrB, 500 nM GreB, 2 mM ATP, 200 μM UTP and 200 μM GTP (continuous-tracking methodology). b, Nanomanipulation time-trace obtained as in a but for negatively supercoiled DNA (−sc). c–f, Lifetime distributions for the Mfd–RNAP intermediate in the presence of UvrA and UvrB as above are essentially independent of both the cause of RNAP stalling (either a C-less cassette or a CPD) and supercoiling of the DNA (positive or negative). For positively supercoiled template the mean lifetime observed using DNA bearing a C-less cassette is 132 ± 7 s (s.e.m., n = 210 (c), see overview in Extended Data Fig. 6d) and for DNA bearing a CPD it is 141 ± 20 s (s.e.m., n = 58 (e)). For negatively supercoiled template the mean lifetime observed using DNA bearing a C-less cassette is 132 ± 13 s (s.e.m., n = 65 (d)) and for DNA bearing a CPD it is 157 ± 65 s (s.e.m., n = 10 (f)).
Extended Data Figure 6 Lifetime distributions of the Mfd–RNAP intermediate as a function of UvrA concentration, using the tethered-DNA assay.
The DNA substrate used in these experiments was positively supercoiled and contained a C-less cassette, and data were collected using the continuous-tracking methodology in the presence of 10–20 pM RNAP holoenzyme, 500 nM GreB, 100 nM Mfd, 2 mM ATP, 200 μM UTP, 200 μM GTP and 250 nM UvrB. The UvrA concentration was (a) 50 pM, (b) 75 pM and (c) 100 pM. Red lines show the result of global fitting to a difference-of-two exponentials characteristic of a Michaelis–Menten process, using the rate-limiting forward catalytic step extracted from classical Michaelian analysis of the mean times (Fig. 2d) as an additional constraint. d, Overview of lifetimes of the Mfd–RNAP complex measured with the tethered-DNA assay and as a function of template supercoiling, cause of RNAP stalling and UvrA concentration, as presented in this paper. UvrB was fixed at 250 nM throughout.
Extended Data Figure 7 Mfd–RNAP-UvrAB control experiments, using the tethered-DNA assay.
These experiments were performed on positively supercoiled DNA substrate (+sc) bearing a C-less cassette and using the standard pulse-chase methodology. a, No ATP control: ATP dependence of UvrAB remodelling of Mfd–RNAP intermediate. Black down-arrows indicate component infusion as follows. RNAP: we first introduce 25 pM RNAP holoenzyme, 500 nM GreB, 200 μM ATP, 200 μM UTP and 200 μM GTP, and wait for RNAP to stall on DNA. Wash: we next wash out all free components except for GreB. Mfd ATP: we next infuse 100 nM Mfd, 500 nM GreB and 50 μM ATP and wait for Mfd to remodel RNAP and form the Mfd–RNAP intermediate. Wash: we next wash out all free components except GreB. UvrAB: we next infuse 50 pM UvrA, 250 nM UvrB and 500 nM GreB. We wait several thousand seconds, without any observed change in the intermediate state. ATP: finally, we infuse 2 mM ATP into the reaction and rapidly observe resolution of the intermediate species. b, No Mfd control: UvrAB does not functionally interact with RNAP in the absence of Mfd. Stalled RNAP formed as in a is not displaced in the presence of (down-arrow) 50 pM UvrA, 250 nM UvrB, 500 nM GreB and 2 mM ATP. c, No UvrA control: lifetime distribution for the Mfd–RNAP intermediate in the presence of UvrB alone. Stalled RNAP is formed as in a. We then wash out free RNAP while maintaining GreB and NTPs in solution, and add 100 nM Mfd, 250 nM UvrB and 2 mM ATP while maintaining GreB and NTPs in solution. The lifetime of the Mfd–RNAP intermediate thus formed remains long-lived (642 ± 22 s (s.e.m.), n = 80) with Gaussian statistics.
Extended Data Figure 8 Control experiments for UvrA and UvrB interactions with DNA in the absence of damage as seen in the tethered-DNA supercoiling assay.
Experiments were conducted on positively supercoiled DNA bearing a CTP-less cassette. a, UvrA alone compacts undamaged, supercoiled DNA in a non-specific manner even at concentrations as low as 10 pM. Trace shown obtained with 1 mM ATP; the same phenomenon is observed in the absence of ATP (data not shown). b, UvrB prevents non-specific interaction of UvrA with DNA; (t = 0 s) 250 nM UvrB alone does not compact DNA, although it transiently interacts non-specifically and briefly with DNA in the presence of 1 mM ATP (see c–f). The same phenomenon is observed in the absence of ATP (data not shown); (t = 2,000 s) addition of UvrB also prevents UvrA from compacting DNA non-specifically. On the basis of these data we set the working UvrB concentration to 250 nM: our measurements with UvrA can thus go up to 100 pM, which remains more than 90% saturated by this concentration of UvrB as shown by the fact that we can perform measurements without DNA compaction. c, d, Time-traces obtained on positively supercoiled DNA in the presence of 250 nM UvrB and 1 mM ATP show supercoiling-dependence of the dwell time (tdwell) of UvrB-DNA ‘wrapping’ events. Indeed the amplitude of these events (~50–100 nm) is consistent with titration of a large positive supercoil by formation of a tight/compact, positive wrap of DNA around UvrB as observed in AFM imaging27. e, f, Histograms of the dwell time of the wrap state obtained above are fitted to single-exponential distributions, with a mean dwell time of (e) 28 ± 2 s (s.e.m., n = 175, +5 turns), and (f) 66 ± 5 s (s.e.m., n = 117, +6 turns). By performing experiments with no more than 250 nM UvrB and with only +4 turns of positive supercoiling, this wrap state is of order 10 s and does not significantly interfere with detection of Mfd–RNAP intermediates or their resolution, and UvrB safely inhibits DNA compaction activity by UvrA.
Extended Data Figure 9 Correlation between resolution of the Mfd–RNAP intermediate and loss of fluorescent signal from labelled RNAP or Mfd in the NanoCOSM assay.
We plot the time elapsed between loss of fluorescence signal from (a) fluorescent RNAP or (b) fluorescent Mfd and nanomechanical resolution of the Mfd–RNAP intermediate as observed in the magnetic trap, as shown in Fig. 3a, b. In both cases the vast majority of events are correlated as shown by the fact that loss of fluorescence and nanomechanical resolution of the intermediate temporally coincide (that is, the time between the two events is nil). Loss of fluorescence before nanomechanical resolution (that is, indicated as positive times) is most probably due to spontaneous photobleaching of the DY-549 fluorophore used to label proteins. No significant difference is observed between DNA substrates bearing a CPD or bearing a C-less cassette.
Extended Data Figure 10 Characterization of specific, control and non-specific GGR incision using the tethered-DNA assay.
a, b, Time-traces showing GGR incision on positively and negatively supercoiled CPD-bearing DNA. For positively supercoiled DNA (+sc), addition of UvrABC proteins (1 nM UvrA, 250 nM UvrB, 100 pM UvrC and 100 pM pUC18 competitor DNA) and 1 mM ATP led to DNA incision and an abrupt loss of supercoiling. The average GGR incision times are 1,230 ± 195 s (s.e.m., n = 72 events) and 1,156 ± 256 s (s.e.m., n = 40 events) for +sc and −sc, respectively; see Fig. 3f, g for distributions and fits. c, As in a, but in the absence of ATP. The absence of incision was confirmed on 22 molecules over a ~4 h window. Upon supplementing the reactions with 1 mM ATP (red down-arrow) incision rapidly takes place. d, UvrC (100 pM) and ATP (1 mM) are unable to incise positively supercoiled, CPD-bearing DNA. The absence of incision was confirmed on 31 molecules over a ~2 h window. e, Incision times for UvrABC (as above) acting on positively supercoiled DNA bearing a C-less cassette (that is, undamaged) are essentially normally distributed (red line) with a mean of 2,922 ± 222 s (n = 44 events; the fit was obtained by excluding points between 3,000 and 4,000 s). f, Incision times for UvrABC (as above) acting on negatively supercoiled DNA bearing a C-less cassette are essentially normally distributed with a mean of 2,471 ± 377 s (s.e.m., n = 28 events; the fit was obtained by excluding points below 1,000 s). g, Incision times for UvrABC acting on positively supercoiled DNA bearing a CPD protected by stalled RNAP in the absence of Mfd are essentially normally distributed with a mean of 2,348 ± 672 s (s.e.m., n = 31 events). Red lines are guides to the eye (c, d) and overall results confirm all of UvrAB, UvrC and ATP are required for GGR incision. Results from e–g further indicate that non-specific DNA incision by the complete GGR system can take place in this assay; however, it is slow enough to permit measurement of faster specific incision rates discussed in Fig. 3. We propose these incision events are in fact specific to the multiple biotin- and digoxigenin-based tethers at the ends of the DNA construct, which can ultimately be recognized as DNA damage by the GGR machinery9. Because only nicking at the first tethering biotin or dig, and within the 2 kbp fragment, will result in loss of supercoiling, then, statistically, multiple incisions at multiple tethers must be realized before loss of supercoiling, resulting in a normal distribution. This can be compared to DNA incision by endonuclease, the time distribution of which is single-exponential28. As incision on these constructs is significantly slower and obeys different statistics than that on CPD-containing DNA, we conclude that our single-molecule measurements can indeed isolate CPD-specific from non-specific incision by the GGR machinery.
Supplementary information
Supplementary Data
This file contains Supplementary Table 1. (PDF 79 kb)
Rights and permissions
About this article
Cite this article
Fan, J., Leroux-Coyau, M., Savery, N. et al. Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature 536, 234–237 (2016). https://doi.org/10.1038/nature19080
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19080
This article is cited by
-
Shifted PAMs generate DNA overhangs and enhance SpCas9 post-catalytic complex dissociation
Nature Structural & Molecular Biology (2023)
-
Nanomanipulation in Biomedical Applications
Current Robotics Reports (2021)
-
Single-molecule live-cell imaging visualizes parallel pathways of prokaryotic nucleotide excision repair
Nature Communications (2020)
-
Single-molecule imaging reveals molecular coupling between transcription and DNA repair machinery in live cells
Nature Communications (2020)
-
Rho-dependent transcription termination in bacteria recycles RNA polymerases stalled at DNA lesions
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.