Abstract
The non-receptor protein tyrosine phosphatase SHP2, encoded by PTPN11, has an important role in signal transduction downstream of growth factor receptor signalling and was the first reported oncogenic tyrosine phosphatase1. Activating mutations of SHP2 have been associated with developmental pathologies such as Noonan syndrome and are found in multiple cancer types, including leukaemia, lung and breast cancer and neuroblastoma1,2,3,4,5. SHP2 is ubiquitously expressed and regulates cell survival and proliferation primarily through activation of the RAS–ERK signalling pathway2,3. It is also a key mediator of the programmed cell death 1 (PD-1) and B- and T-lymphocyte attenuator (BTLA) immune checkpoint pathways6,7. Reduction of SHP2 activity suppresses tumour cell growth and is a potential target of cancer therapy8,9. Here we report the discovery of a highly potent (IC50 = 0.071 μM), selective and orally bioavailable small-molecule SHP2 inhibitor, SHP099, that stabilizes SHP2 in an auto-inhibited conformation. SHP099 concurrently binds to the interface of the N-terminal SH2, C-terminal SH2, and protein tyrosine phosphatase domains, thus inhibiting SHP2 activity through an allosteric mechanism. SHP099 suppresses RAS–ERK signalling to inhibit the proliferation of receptor-tyrosine-kinase-driven human cancer cells in vitro and is efficacious in mouse tumour xenograft models. Together, these data demonstrate that pharmacological inhibition of SHP2 is a valid therapeutic approach for the treatment of cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Grossmann, K. S., Rosário, M., Birchmeier, C. & Birchmeier, W. The tyrosine phosphatase Shp2 in development and cancer. Adv. Cancer Res. 106, 53–89 (2010)
Chan, R. J. & Feng, G. S. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood 109, 862–867 (2007)
Matozaki, T., Murata, Y., Saito, Y., Okazawa, H. & Ohnishi, H. Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci. 100, 1786–1793 (2009)
Mohi, M. G. & Neel, B. G. The role of Shp2 (PTPN11) in cancer. Curr. Opin. Genet. Dev. 17, 23–30 (2007)
Östman, A., Hellberg, C. & Böhmer, F. D. Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer 6, 307–320 (2006)
Gavrieli, M., Watanabe, N., Loftin, S. K., Murphy, T. L. & Murphy, K. M. Characterization of phosphotyrosine binding motifs in the cytoplasmic domain of B and T lymphocyte attenuator required for association with protein tyrosine phosphatases SHP-1 and SHP-2. Biochem. Biophys. Res. Commun. 312, 1236–1243 (2003)
Okazaki, T., Chikuma, S., Iwai, Y., Fagarasan, S. & Honjo, T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 14, 1212–1218 (2013)
Prahallad, A. et al. PTPN11 is a central node in intrinsic and acquired resistance to targeted cancer drugs. Cell Reports 12, 1978–1985 (2015)
Schneeberger, V. E. et al. Inhibition of Shp2 suppresses mutant EGFR-induced lung tumors in transgenic mouse model of lung adenocarcinoma. Oncotarget 6, 6191–6202 (2015)
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012)
Scott, L. M. et al. Shp2 protein tyrosine phosphatase inhibitor activity of estramustine phosphate and its triterpenoid analogs. Bioorg. Med. Chem. Lett. 21, 730–733 (2011)
Grosskopf, S. et al. Selective inhibitors of the protein tyrosine phosphatase SHP2 block cellular motility and growth of cancer cells in vitro and in vivo. ChemMedChem 10, 815–826 (2015)
He, R. et al. Exploring the existing drug space for novel pTyr mimetic and SHP2 inhibitors. ACS Med. Chem. Lett. 6, 782–786 (2015)
Hellmuth, K. et al. Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proc. Natl Acad. Sci. USA 105, 7275–7280 (2008)
Zeng, L. F. et al. Therapeutic potential of targeting the oncogenic SHP2 phosphatase. J. Med. Chem. 57, 6594–6609 (2014)
Pluskey, S., Wandless, T. J., Walsh, C. T. & Shoelson, S. E. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J. Biol. Chem. 270, 2897–2900 (1995)
Hof, P., Pluskey, S., Dhe-Paganon, S., Eck, M. J. & Shoelson, S. E. Crystal structure of the tyrosine phosphatase SHP-2. Cell 92, 441–450 (1998)
Manley, P. W. et al. Extended kinase profile and properties of the protein kinase inhibitor nilotinib. Biochim. Biophys. Acta. 1804, 445–453 (2010)
Bender, A. et al. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. ChemMedChem 2, 861–873 (2007)
Szczepankiewicz, B. G. et al. Discovery of a potent, selective protein tyrosine phosphatase 1B inhibitor using a linked-fragment strategy. J. Am. Chem. Soc. 125, 4087–4096 (2003)
Gilmartin, A. G. et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat. Chem. Biol. 10, 181–187 (2014)
Hoffman, G. R. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl Acad. Sci. USA 111, 3128–3133 (2014)
Shao, D. D. et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res. 23, 665–678 (2013)
Clare, J. J., Tate, S. N., Nobbs, M. & Romanos, M. A. Voltage-gated sodium channels as therapeutic targets. Drug Discov. Today 5, 506–520 (2000)
Zhao, H. et al. Isoxazole carboxylic acids as protein tyrosine phosphatase 1B (PTP1B) inhibitors. Bioorg. Med. Chem. Lett. 14, 5543–5546 (2004)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Bricogne, G. et al. BUSTER version 2.8.0. (Global Phasing Ltd., 2009)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Weisberg, E. et al. Inhibition of wild-type p53-expressing AML by the novel small molecule HDM2 inhibitor CGM097. Mol. Cancer Ther. 14, 2249–2259 (2015)
Acknowledgements
Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract number DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
Y.P.C., F.F., H.-X.H., K.H., S.L., J.S., P.Z., H.M.C. performed or directed cellular assays data generation and analysis; P.F., M.G.A., Z.B.K., S.H., E.P., C.Q., S.S, P.W., J.-H.Z. and P.D.F. performed or directed biochemical experiments; B.A., V.G.C., B.F., H.G., L.R.L.B., M.J.M., M.P., G.Y. and J.Y. performed or directed in vivo pharmacology or pharmacokinetic/pharmacodynamics experiments and data analysis; J.R.D. and K.V. directed or performed bioinformatics analyses; M.J.L., J.G.-F., C.F., C.H.-T.C, Z.C., D.G., R.K., M.K., J.L., F.L., G.L., D.M., M.P., L.P., M.D.S., T.S., S.W. Designed, synthesized and/or directed the design or synthesis of SHP2 inhibitors; Z.D., M.K., and S.W. performed protein and inhibitor structural modelling or cheminformatics analyses; M.F., J.J. and T.S. designed, directed or performed biophysics experiments; M.F. and T.S. directed or performed x-ray crystallography experiments; Y.P.C., J.R.D., L.R.L.B., M.F., M.J.M., K.V., H.M.C., T.S., W.R.S. and P.D.F. prepared figures and tables for the main text and Supplementary Information; Y.P.C., M.J.L., J.R.D., L.R.L.B., M.J.M., K.V., N.J.K., H.M.C., T.S., W.R.S. and P.D.F. wrote and edited the main text and Supplementary Information; P.D.F., N.J.K., T.R., T.S., W.R.S. and M.W. contributed to overall project oversight.
Corresponding authors
Ethics declarations
Competing interests
All authors performed the work herein as employees of the Novartis Institutes for Biomedical Research, Inc.
Additional information
Reviewer Information Nature thanks B. Neel and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
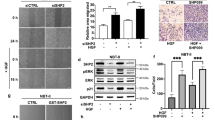
Extended Data Figure 1 SHP2 depletion inhibits the growth of RTK-amplified cancer cells.
a, Cells expressing dox-inducible SHP2 shRNA in various RTK-amplified cancer cells were generated including SUM52 (FGFR2), KATOIII (FGFR2), MDA-MB-468, KYSE520 (EGFR), NCI-H2190 (HER2), NCI-H2228 (EML4-ALK). b, Stable clones of MDA-MB-231 (KRASG13D) and A2058 BRAFV600E) cancer cells were established as controls. Cells were treated with dox, and colony formation was measured after 11 days by crystal violet staining. c, Western blot showing the expression of SHP2, p-ERK and ERK in the presence (+) or absence (−) of dox in MDA-MB-468 SHP2-depleted cells stably expressing either GFP, wild-type HA–SHP2 or HA–SHP2C459S. d, Western blot of SHP2, p-ERK and ERK in the presence (+) or absence (−) of dox in SHP2-depleted SUM52 cells expressing vector control or HA–KRASG12V. In c and d, dox treatment induces depletion of endogenous SHP2 protein and simultaneously expression of the exogenous proteins GFP, wild-type HA–SHP2, HA–SHP2C459S or HA–KRASG12V.
Extended Data Figure 2 Phosphatase activity is required for cancer growth.
a, Western blot of SHP2, p-ERK and ERK in SHP2-depleted SUM52 cells stably expressing vehicle, wild-type SHP2 or HA–SHP2C459S. Note, four lanes corresponding to an unrelated study were removed from the image. All lanes originated from the same gel at the same exposure. b, Colony formation of SHP2-depleted SUM52 cells stably expressing vehicle, wild-type SHP2 or HA–SHP2C459S. SHP2 knockdown and SHP2 variant re-expression perform using dox treatment. Colony formation was monitored after 11 days with crystal violet staining. c, Phosphatase activity of SHP2-depleted SUM52 cells stably expressing HA–SHP2 or HA–SHP2C459S. Cells were treated with dox for 3 days. SHP2 protein was immunoprecipitated from cell lysates and phosphatase activity was measured using DiFMUP assay. Data are presented as mean ± s.d. (n = 3).
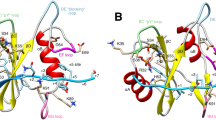
Extended Data Figure 3 Thermodynamic characterization of the SHP099–SHP2 binding complex, comparison of SHP2 and SHP1’s allosteric pocket and characterization of SHP099-resistant SHP2 mutants.
a, Isothermal titration calorimetry of SHP099 binding to SHP2. SHP099 binds stoichiometrically to SHP2 with a dissociation constant measured at 73 ± 15 nM. b, Structural differences in the central tunnel between SHP1 and SHP2. Ribbon representation of SHP2 (multi-colour) and SHP1 (grey) X-ray structures in the closed conformation. The PTP (tan) and N-SH2 (green) domain overlay well (r.m.s.d. < 1.5 Å), however the C-SH2 (blue) domain has a significantly different orientation. c, Surface representation of SHP2–SHP099 co-crystal structure. d, Surface representation of SHP1 with SHP099 modelled on the basis of the SHP2 superimposition. Central tunnel is significantly larger in SHP1 owing to a change in orientation of the C-SH2 domain. This change repositions the linker between the two SH2 domains removing several key interactions, highlighted by residue Arg109 in SHP1 and by Arg111 in SHP2 (equivalent residues). e, Biochemical activity of wild-type SHP2, SHP2Q257L and SHP2T253M/Q257L. SHP2 activity was determined using DiFMUP in the presence of various concentrations of 2P-IRS-1. Data points along the line represent the mean of two replicate values. SHP2Q257L and SHP2T253M/Q257L retain activity regulation and 2P-IRS-1 activation potential comparable to wild-type SHP2 but are 18- and <1,000-fold less sensitive to SHP099 inhibition.
Extended Data Figure 4 Cellular activity of SHP099.
a, Western blot of SHP2, p-ERK, ERK and p-AKT from KYSE520, MDA-MB-468 or A2058 cells treated with SHP099 (1, 3, 10 μM). Note, three lanes corresponding to an unrelated study were removed from the image. All lanes originated from the same gel at the same exposure. b, Colony formation of KYSE520, MDA-MB-468, and A2058 in the presence of SHP099. Colony formation was measured after 11 days of SHP099 treatment by crystal violet staining. c, SHP099 inhibitory activity against cell lines KYSE520 (EGFR-amplified), MV-411(FLT3-ITD), MOLM-13 (FLT3-ITD), Kasumi (c-Kit altered) and negative control A2058 (BRAFV600E) treated with SHP099 concentration varying from 0.0046 to 20 μM. Cellular viability was measured using CellTiter-Glo. Data presented as mean ± s.d. (n = 3). d, Comparison of SHP099 activity with Lapatinib in a panel of 26 colorectal cell lines. SHP099 sensitivity correlates with sensitivity to Lapatinib, a potent tyrosine kinase inhibitor against Her1/2 and EGFR. RAS- and BRAF-mutated cell lines are shown in blue. Cellular viability was measured using CellTiter-Glo. The corresponding data and cell line genotypes are included in Supplementary Information Table 2.
Extended Data Figure 5 SHP2 depletion or inhibition by SHP099 assessed in in vivo xenograft models.
a, Western blot of SHP2 and GAPDH in KYSE520 xenograft lysates following 14 days of dox treatment (+). b, Response of p-ERK in tumour xenograft lysates following 14 days of dox treatment. c, Antitumour efficacy of SHP099 administered orally for 14 consecutive days at the doses and schedules indicated. Data are plotted as the treatment mean ± s.e.m. (n = 9). d, Body weight of mice bearing subcutaneous KYSE520 xenografts and administered either SHP099 (100 mg per kg daily), erlotinib (80 mg per kg daily) or vehicle for 14 consecutive days. Data are presented as treatment mean ± s.e.m. (n = 7). e, Body weight of mice bearing orthotopic primary tumour derived xenografts and administered an oral gavage of SHP099 at 75 mg per kg daily. Data are presented as treatment mean ± s.e.m. (n = 7). f, The mouse spleen weight measurement of mice bearing the orthotopic AML xenograft model following 34 days of once-daily dosing with SHP099 at 75 mg per kg. Data are presented as treatment mean ± s.e.m. (n = 7).
Supplementary information
Supplementary Figure 1
This file contains the raw data for Figure 3e and Extended Data Figures 1a-d, 2a, 4a and 5a. (PDF 1634 kb)
Supplementary Table 1
This file shows the SHP099 inhibitory activity against hematopoietic cell lines. Data used to generate Figure 3b. (XLSX 10 kb)
Supplementary Table 2
This file shows the Inhibition of colorectal cancer cell lines proliferation by SHP099 or Lapatinib. Data used to generate extended data figure 4d. (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Chen, YN., LaMarche, M., Chan, H. et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535, 148–152 (2016). https://doi.org/10.1038/nature18621
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18621
This article is cited by
-
Phosphoproteomic investigation of targets of protein phosphatases in EGFR signaling
Scientific Reports (2024)
-
From bench to bedside: current development and emerging trend of KRAS-targeted therapy
Acta Pharmacologica Sinica (2024)
-
Targeting protein phosphatases in cancer immunotherapy and autoimmune disorders
Nature Reviews Drug Discovery (2023)
-
Mapping the landscape of genetic dependencies in chordoma
Nature Communications (2023)
-
SHP-2 and PD-1-SHP-2 signaling regulate myeloid cell differentiation and antitumor responses
Nature Immunology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.