Abstract
Viral proteins mimic host protein structure and function to redirect cellular processes and subvert innate defenses1. Small basic proteins compact and regulate both viral and cellular DNA genomes. Nucleosomes are the repeating units of cellular chromatin and play an important part in innate immune responses2. Viral-encoded core basic proteins compact viral genomes, but their impact on host chromatin structure and function remains unexplored. Adenoviruses encode a highly basic protein called protein VII that resembles cellular histones3. Although protein VII binds viral DNA and is incorporated with viral genomes into virus particles4,5, it is unknown whether protein VII affects cellular chromatin. Here we show that protein VII alters cellular chromatin, leading us to hypothesize that this has an impact on antiviral responses during adenovirus infection in human cells. We find that protein VII forms complexes with nucleosomes and limits DNA accessibility. We identified post-translational modifications on protein VII that are responsible for chromatin localization. Furthermore, proteomic analysis demonstrated that protein VII is sufficient to alter the protein composition of host chromatin. We found that protein VII is necessary and sufficient for retention in the chromatin of members of the high-mobility-group protein B family (HMGB1, HMGB2 and HMGB3). HMGB1 is actively released in response to inflammatory stimuli and functions as a danger signal to activate immune responses6,7. We showed that protein VII can directly bind HMGB1 in vitro and further demonstrated that protein VII expression in mouse lungs is sufficient to decrease inflammation-induced HMGB1 content and neutrophil recruitment in the bronchoalveolar lavage fluid. Together, our in vitro and in vivo results show that protein VII sequesters HMGB1 and can prevent its release. This study uncovers a viral strategy in which nucleosome binding is exploited to control extracellular immune signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Elde, N. C. & Malik, H. S. The evolutionary conundrum of pathogen mimicry. Nature Rev. Microbiol. 7, 787–797 (2009)
Smale, S. T., Tarakhovsky, A. & Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 32, 489–511 (2014)
Lischwe, M. A. & Sung, M. T. A histone-like protein from adenovirus chromatin. Nature 267, 552–554 (1977)
Chatterjee, P. K., Vayda, M. E. & Flint, S. J. Adenoviral protein VII packages intracellular viral DNA throughout the early phase of infection. EMBO J. 5, 1633–1644 (1986)
Vayda, M. E., Rogers, A. E. & Flint, S. J. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 11, 441–460 (1983)
Kang, R. et al. HMGB1 in health and disease. Mol. Aspects Med. 40, 1–116 (2014)
Lotze, M. T. & Tracey, K. J. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nature Rev. Immunol. 5, 331–342 (2005)
Paschos, K. & Allday, M. J. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 18, 439–447 (2010)
Marazzi, I. et al. Suppression of the antiviral response by an influenza histone mimic. Nature 483, 428–433 (2012)
Ferrari, R., Berk, A. J. & Kurdistani, S. K. Viral manipulation of the host epigenome for oncogenic transformation. Nature Rev. Genet. 10, 290–294 (2009)
Knipe, D. M. et al. Snapshots: chromatin control of viral infection. Virology 435, 141–156 (2013)
Ferrari, R. et al. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16, 663–676 (2014)
Wykes, S. M. & Krawetz, S. A. The structural organization of sperm chromatin. J. Biol. Chem. 278, 29471–29477 (2003)
Lin, S. & Garcia, B. A. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 512, 3–28 (2012)
Shechter, D., Dormann, H. L., Allis, C. D. & Hake, S. B. Extraction, purification and analysis of histones. Nature Protocols 2, 1445–1457 (2007)
Teves, S. S. & Henikoff, S. Salt fractionation of nucleosomes for genome-wide profiling. Methods Mol. Biol. 833, 421–432 (2012)
Falk, S. J. et al. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703 (2015)
White, A. E., Hieb, A. R. & Luger, K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci. Rep. 6, 19122 (2016)
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Fedor, M. J. & Daniell, E. Acetylation of histone-like proteins of adenovirus type 5. J. Virol. 35, 637–643 (1980)
Robinson, C. M. et al. Molecular evolution of human adenoviruses. Sci. Rep. 3, 1812 (2013)
Gyurcsik, B., Haruki, H., Takahashi, T., Mihara, H. & Nagata, K. Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry 45, 303–313 (2006)
Haruki, H., Okuwaki, M., Miyagishi, M., Taira, K. & Nagata, K. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 80, 794–801 (2006)
Scaffidi, P., Misteli, T. & Bianchi, M. E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418, 191–195 (2002)
Sapojnikova, N. et al. Biochemical observation of the rapid mobility of nuclear HMGB1. Biochim. Biophys. Acta 1729, 57–63 (2005)
Lu, B. et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488, 670–674 (2012)
Koziol-White, C. J., Damera, G. & Panettieri, R. A. Jr. Targeting airway smooth muscle in airways diseases: an old concept with new twists. Expert Rev. Respir. Med. 5, 767–777 (2011)
Ueno, H. et al. Contributions of high mobility group box protein in experimental and clinical acute lung injury. Am. J. Respir. Crit. Care Med. 170, 1310–1316 (2004)
Johnson, J. S. et al. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J. Virol. 78, 6459–6468 (2004)
Karen, K. A. & Hearing, P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 85, 4135–4142 (2011)
Khandelia, P., Yap, K. & Makeyev, E. V. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl Acad. Sci. USA 108, 12799–12804 (2011)
Kozarsky, K. F., Jooss, K., Donahee, M., Strauss, J. F., III & Wilson, J. M. Effective treatment of familial hypercholesterolaemia in the mouse model using adenovirus-mediated transfer of the VLDL receptor gene. Nature Genet. 13, 54–62 (1996)
Orazio, N. I., Naeger, C. M., Karlseder, J. & Weitzman, M. D. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J. Virol. 85, 1887–1892 (2011)
Le, L. P. et al. Core labeling of adenovirus with EGFP. Virology 351, 291–302 (2006)
Reich, N. C., Sarnow, P., Duprey, E. & Levine, A. J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128, 480–484 (1983)
Lilley, C. E., Chaurushiya, M. S., Boutell, C., Everett, R. D. & Weitzman, M. D. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 7, e1002084 (2011)
Das, S., MacDonald, K., Chang, H.-Y. S. & Mitzner, W. A simple method of mouse lung intubation. J. Vis. Exp . 73, e50318 (2013)
Jeyaseelan, S., Chu, H. W., Young, S. K. & Worthen, G. S. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect. Immun. 72, 7247–7256 (2004)
Nick, J. A. et al. Role of p38 mitogen-activated protein kinase in a murine model of pulmonary inflammation. J. Immunol. 164, 2151–2159 (2000)
Zaret, K. Micrococcal nuclease analysis of chromatin structure. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21.1 (2005)
Wessel, D. & Flügge, U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984)
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 (2008)
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011)
Tanaka, Y. et al. Expression and purification of recombinant human histones. Methods 33, 3–11 (2004)
Luger, K., Rechsteiner, T. J., Flaus, A. J., Waye, M. M. & Richmond, T. J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 (1997)
Hasson, D. et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nature Struct. Mol. Biol . 20, 687–695 (2013)
Sekulic, N., Bassett, E. A., Rogers, D. J. & Black, B. E. The structure of (CENP-A–H4)2 reveals physical features that mark centromeres. Nature 467, 347–351 (2010)
Kulej, K., Avgousti, D. C., Weitzman, M. D. & Garcia, B. A. Characterization of histone post-translational modifications during virus infection using mass spectrometry-based proteomics. Methods 90, 8–20 (2015)
Cooper, P. R. & Panettieri, R. A. Jr. Steroids completely reverse albuterol-induced β2-adrenergic receptor tolerance in human small airways. J. Allergy Clin. Immunol. 122, 734–740 (2008)
Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002)
Blumenthal, D., Goldstien, L., Edidin, M. & Gheber, L. A. Universal approach to FRAP analysis of arbitrary bleaching patterns. Sci. Rep. 5, 11655 (2015)
Acknowledgements
We thank members of the Weitzman laboratory for insightful discussions and input, especially R. Dilley and B. Simpson for generating reagents. We also thank R. Panetierri and C. Koziol-White for providing precision-cut lung slices. We are grateful to D. Curiel for sharing recombinant protein-VII–GFP vectors and L. Gerace for anti-protein-VII antibodies. We thank the Penn Vector Core for assistance in purifying recombinant vectors, the Penn CDB Microscopy Core for imaging and FRAP assistance, and the CHOP Pathology core for immunostaining of mouse lungs. We thank members of the Black, Garcia and Worthen laboratories for technical help. We thank C. Bassing, I. Brodsky, J. Henao-Mejia, R. Kohli, C. Lopez, A. Resnick, S. Shin, K. Spindler and J. Weitzman for advice and critical reading of the manuscript. D.C.A. was supported in part by T32 CA115299 and F32 GM112414. N.J.P. was supported in part by T32 NS007180. N.S. was supported in part by funding from the American Cancer Society. Research was supported by grants from the National Institutes of Health (CA097093 to M.D.W., AI102577 and CA122677 to P.H., AI118891 and GM110174 to B.A.G., and GM082989 to B.E.B.), the Institute for Immunology of the University of Pennsylvania, and funds from the Children’s Hospital of Philadelphia (M.D.W.).
Author information
Authors and Affiliations
Contributions
D.C.A. and M.D.W. conceived the project and designed experiments; D.C.A., C.H., N.S., J.P., N.J.P. and E.D.R. performed the experiments; D.C.A., C.H. and J.P. generated constructs and cell lines; K.K., R.C.M., S.H.S. and B.A.G. performed MS analysis; P.O. and P.H. generated Ad5-flox-VII virus and provided 293-Cre cell line; D.C.A. and D.B. performed the FRAP experiments; A.J.P. and G.S.W. conducted all mouse experiments; B.E.B. and B.A.G. designed experiments and interpreted the data; D.C.A. and M.D.W. interpreted the data and wrote the manuscript and all authors were involved in editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
All proteomic raw files have been deposited in the Chorus database under project number 1047 (https://chorusproject.org/).
Reviewer Information Nature thanks M. Bianchi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
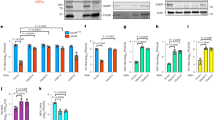
Extended Data Figure 1 Adenovirus protein VII distorts chromatin.
a, Protein VII localizes to cellular chromatin and viral replication centres in U2OS cells similarly to SAECs in Fig. 1a. b, Protein VII messenger RNA levels measured by quantitative PCR showing that after 4 days of induction in the A549 cell line, the level of protein VII transcripts is approximately 10% of that measured during infection at 16 hpi. Despite the low relative level, this amount of protein VII is sufficient to cause dramatic changes in the nucleus (graph shows mean ± s.d., n = 3 biological replicates). c, Inducible cell lines of U2OS and HeLa expressing protein-VII–HA show chromatin localization and distortion, similar to A549 cells in Fig. 1c. d, Inducible A549 cell lines expressing viral protein V, the precursor for protein VII (preVII) or cellular protamine PRM1 with C-terminal HA tags. Although all three proteins possess a large number of charged residues, none are sufficient to distort cellular chromatin or increase nuclear size as observed with mature protein VII. Scale bars, 10 μm.
Extended Data Figure 2 Protein VII associates tightly with chromatin and binds DNA and nucleosomes in vitro.
a, Western blot analysis showing protein VII in histone extracts from infected HeLa cells at 24 hpi. b, Chromatin fractionation of lysates from A549 cells that were uninfected (mock) or infected for 24 h with Ad5. Viral and cellular proteins were detected by western blotting with various antibodies as indicated. c, Agarose gel analysis of DNA extracted from nuclear fractionation experiments, indicating that the size of DNA is between 100 and 200 bp and elutes predominantly in the higher-salt fractions. d, Chromatin fractionation of cells induced to express protein VII, indicating that protein VII is present in the highest-salt fraction from the first day of induction. e, f, Recombinant protein-VII–His binds DNA. Incubating increasing molar amounts of protein VII with 195 bp DNA results in shifts by native gel electrophoresis, indicating protein-VII–DNA complex formation. Staining with either ethidium bromide (e) or Coomassie (f) are shown to verify the presence of DNA and protein, respectively. g, Ethidium bromide staining shows DNA content of nucleosome shifts from gel in Fig. 1f.
Extended Data Figure 3 Bioanalyzer examination of MNase-digested nucleosomes and protein-VII–nucleosome complexes.
a, 195 bp nucleosomes or protein-VII–nucleosome complexes were incubated with MNase for the indicated times, the reaction was stopped, DNA was extracted and analysed. As in Fig. 1g, nucleosomes are shown in black and protein-VII–nucleosome complexes in orange. The presence of protein VII pauses digestion at 165 bp, suggesting that protein VII is blocking access to the DNA. b, 147 bp nucleosomes or protein-VII–nucleosome complexes were incubated with MNase for the indicated times, the reaction was stopped, DNA was extracted and analysed. Graphs show nucleosomes in grey and protein-VII–nucleosome complexes in orange. The presence of protein VII completely blocks digestion even after nucleosomes alone have been digested well beyond the core particle. In contrast to what would be expected for linker histones, protein VII protects the core nucleosome particle from digestion. These data indicate that protein VII may be masking the substrate for MNase through complex formation. This represents a unique mechanism of nucleosome binding and suggests a model for blocking DNA access in cellular chromatin during infection.
Extended Data Figure 4 Purification of protein VII from infected cells.
a, Coomassie-stained SDS–PAGE analysis of fractions from RP-HPLC in Fig. 2a. The bands in fraction 38–41 min correspond to histone H1. Protein VII and V, as indicated, were verified by MS analysis (data not shown). The slight upward shift of the protein VII bands in the later peak corresponds to the higher abundance of protein preVII, as seen by HPLC in Fig. 2a. b, Western blot analysis of protein VII in HPLC fractions from a. c, Time course of infection followed by histone extraction and HPLC analysis. MS analysis verified peaks in each sample as indicated.
Extended Data Figure 5 Representative mass spectra.
a–f, Annotated MS/MS spectra of identified peptides of protein VII containing PTMs (a–c, acetylated peptides; d–f, phosphorylated peptides). The images represent the observed fragment ions collected using MS/MS collision-induced dissociation (CID). Coloured lines represent matches between observed and expected fragment ions of the given peptides. Specifically, green lines represent not fragmented precursor mass, blue lines represent matches with y-type fragments, red lines with b-type fragments, and yellow boxed masses represent fragments containing PTM neutral losses (for example, ions that lost the phosphorylation during fragmentation).
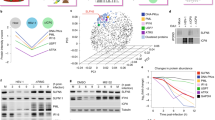
Extended Data Figure 6 Acetylated protein VII spectra from virus particles and analysis of total histone PTM changes upon protein VII expression.
a, Liquid chromatography-mass spectrometry (LC-MS) analysis of unmodified and modified chymotryptic peptide AKKRSDQHPVRVRGHY. On the left, nano-LC-MS-extracted ion chromatograms of protein VII peptides identified in the histone extracts of adenovirus infected cells (Inf) or viral particles (VP). The top left represents the modified form, while the bottom left represents the unmodified form. Non-modified forms were detected in both conditions for Inf and VP, while the acetylated form was unique for the infected sample only (Inf). On the right, full MS spectrum of the modified (top) and unmodified (bottom) peptide. Circled mass represents the monoisotopic signal of the peptide. b, Summary of post-translational modifications detected on protein VII. Peptides shown were identified during infection at various time points with the mature protein VII in the top row and preVII in the bottom row. The numbers in brackets for preVII indicate the location of the same moiety in mature protein VII. Acetylation sites were detected in approximately 3% of peptides for mature protein VII and 2% of peptides in preVII. Phosphorylation was detected in approximately 1% of peptides for mature protein VII and preVII. c, d, Quantification of histone H3 (c) and H4 (d) PTMs in protein-VII–HA-induced (+dox) and -uninduced (−dox) A549 cells from the analysis of crude histone mixtures (n = 3 biological replicates). Positions of PTMs are listed along the x axis. Modification type is indicated by colour as shown. y Axis represents the cumulative extent of PTMs relative to the total histone H3 or H4, respectively. e, Breakdown of the histone marks (H3K14ac, H3K27me1, H3K36me3, H4K20me1, H4K20me2 and H4K20me3) found to be significantly different (n = 3 biological replicates) in terms of relative abundance between the protein-VII–HA-induced and -uninduced states (<5% homoscedastic two-tailed t-test). Mean ± s.d.
Extended Data Figure 7 Bioinformatic analysis of proteins enriched in the high-salt fraction upon protein VII expression.
a, Venn diagram showing overlap between three biological replicates of high-salt-fraction proteins significantly enriched compared with uninduced cells. b, Proteins found significantly enriched in the protein-VII–HA-induced state compared with uninduced (<5% homoscedastic t-test) in all three biological replicates (‘VII–HA induced’ indicates proteins identified only in protein-VII–HA-induced condition). c, d, Classification of proteins significantly enriched in minimum two out of three biological replicates (protein-VII–HA-induced versus uninduced) according to process network enrichment and Gene Ontology biological process (GeneGo MetaCore pathways analysis package; false discovery rate (FDR) < 5%); each Gene Ontology term was ranked using P-value enrichment.
Extended Data Figure 8 Protein VII retains HMGB1 and HMGB2 in chromatin.
a, Western blot of adenovirus-infected or doxycycline-treated A549 cells showing the relative levels of protein VII expression. HMGB1 levels do not change upon infection or protein VII expression. Tubulin is shown as a loading control. b, Quantitative PCR analysis of mRNA transcripts of HMGB1 in various cell types as indicated (for A549, n = 3 biological replicates; for THP-1, n = 2 biological replicates; mean ± s.d.). The levels of HMGB1 do not significantly change. c, Immunofluorescence analysis of a time course of protein-VII–HA (red) induction shown with HMGB1 (green) and DAPI (grey, blue in merge) in A549 cells. Expression of protein-VII–HA results in a change to the HMGB1 distribution upon expression. d, HMGB1 (green) localization changes between 12 and 24 hpi of wild-type adenovirus in A549 cells, and adopts a pattern similar to protein VII as in Fig. 1a. DBP (red) is shown as a marker of infection, DNA is stained with DAPI (blue in merge). e, Same as d showing that HMGB2 adopts the same pattern as HMGB1 during Ad5 infection at 24 hpi. f, Multiple cells showing the same pattern of HMGB1 relocalization upon expressing protein-VII–GFP as in Fig. 3g. g, HMGB1 retention in the high-salt fraction is conserved across adenovirus serotypes. Western blot analysis of HMGB1 from salt-fractionated A549 cells infected with Ad5, Ad9 or Ad12 as shown. Scale bars, 10 μm.
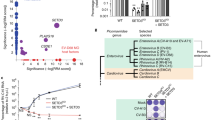
Extended Data Figure 9 Protein VII is necessary and sufficient for chromatin retention of HMGB1 in human and mouse cells.
a, b, Replication of Ad5-flox-VII virus on 293 or 293-Cre cells. Quantitative PCR analysis of viral genomic DNA over a time course of infection (a) shows the DBP gene is increasing exponentially in 293 and 293-Cre cells when infected with Ad5-flox-VII virus. In contrast, PCR for the protein VII gene (b) demonstrates deletion in 293-Cre cells (n = 2 biological replicates, mean ± s.d.). c, Salt fractionation of 293-Cre cells infected with wild-type Ad5, indicating that the Cre recombinase does not interfere with the ability of protein VII to retain HMGB1 in the high-salt chromatin fraction. Protein VII is also necessary for the chromatin retention of HMGB2. d, THP-1 cells transduced to express protein-VII–GFP results in chromatin distortion and HMGB1 retention in chromatin. Immunofluorescence of transduced PMA-treated THP-1 cells showing protein-VII–GFP (green), HMGB1 (red) and DNA (grey, blue in merge). e, Transduction to express protein-VII–GFP is sufficient to relocalize mouse HMGB1 in mouse embryonic fibroblast (MEF) cells. f, Salt fractionation of mouse embryonic fibroblast cells transduced to express protein-VII–GFP. Human Ad5 protein VII is sufficient to retain mouse HMGB1 in the high-salt fraction in MEF cells. The control vector expressing GFP alone does not have this effect.
Extended Data Figure 10 Transduction of mouse lungs demonstrating expression of GFP or protein-VII–GFP.
a, Sections of mouse lungs transduced to express protein-VII–GFP or GFP co-stained for HMGB1. GFP signal shows multiple cell types transduced in both cases. Protein-VII–GFP has a more distinct nuclear signal than GFP, which also appears cytoplasmic. Two sections for each condition are shown to indicate transduction efficiency. b, Same as a but co-stained for prosurfactant-C to mark type II pneumocytes. Some cells are positive for both, confirming that multiple cell types were transduced. c, Zoomed images of individual epithelial cells from mouse lungs showing the characteristic protein-VII–GFP pattern colocalizing with DAPI in the nucleus. GFP only is mostly cytoplasmic. d, Schematic summarizing function of protein VII during infection. Newly synthesized protein VII late during infection can be post-translationally modified and binds to HMGB1, sequestering it on the cellular chromatin and preventing its release. Unmodified protein VII is packaged in viral progeny.
Supplementary information
Supplementary Figures
This file contains the Source data gels for Figures 1d, e, f, 2a, 3b, c, j, k, 4b and Extended Data Figures 2a, b, c, d, e, f, g, 4a, b, c, 8a, g, 9c, f. (PDF 12953 kb)
Supplementary Table 1
Summary of post-translational modifications found on histones upon expression of protein VII. (XLSX 14 kb)
Supplementary Table 2
Total list of proteins significantly changed upon protein VII expression identified by mass spectrometry analysis of high salt fractions. Table includes the log2 fold change of MaxQuant-derived iBAQ values obtained for protein VII-HA induced and uninduced highest salt fractions (600mM). Proteins with homoscedastic two-tailed t-test p-value smaller than 0.05 were considered as significantly altered between the two tested conditions. N/A defines not assigned t-test p-values; this is either due to the presence of the protein in only one condition, or if in one condition the protein was quantified in only one replicate. The number of peptides used for quantification was also highlighted. Commonly occurring contaminants, such as human keratins or trypsin, were removed from the final list. (XLSX 715 kb)
Rights and permissions
About this article
Cite this article
Avgousti, D., Herrmann, C., Kulej, K. et al. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 535, 173–177 (2016). https://doi.org/10.1038/nature18317
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18317
This article is cited by
-
The multifunctional protein HMGB1: 50 years of discovery
Nature Reviews Immunology (2023)
-
The NF-κB Pathway: a Focus on Inflammatory Responses in Spinal Cord Injury
Molecular Neurobiology (2023)
-
Molecular Correlates of Hemorrhage and Edema Volumes Following Human Intracerebral Hemorrhage Implicate Inflammation, Autophagy, mRNA Splicing, and T Cell Receptor Signaling
Translational Stroke Research (2021)
-
MiR-200b in heme oxygenase-1-modified bone marrow mesenchymal stem cell-derived exosomes alleviates inflammatory injury of intestinal epithelial cells by targeting high mobility group box 3
Cell Death & Disease (2020)
-
Hepatitis Delta Virus histone mimicry drives the recruitment of chromatin remodelers for viral RNA replication
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.