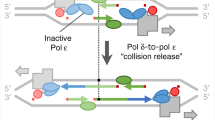
Abstract
The DNA replication and transcription machineries share a common DNA template and thus can collide with each other co-directionally or head-on1,2. Replication–transcription collisions can cause replication fork arrest, premature transcription termination, DNA breaks, and recombination intermediates threatening genome integrity1,2,3,4,5,6,7,8,9,10. Collisions may also trigger mutations, which are major contributors to genetic disease and evolution5,7,11. However, the nature and mechanisms of collision-induced mutagenesis remain poorly understood. Here we reveal the genetic consequences of replication–transcription collisions in actively dividing bacteria to be two classes of mutations: duplications/deletions and base substitutions in promoters. Both signatures are highly deleterious but are distinct from the previously well-characterized base substitutions in the coding sequence. Duplications/deletions are probably caused by replication stalling events that are triggered by collisions; their distribution patterns are consistent with where the fork first encounters a transcription complex upon entering a transcription unit. Promoter substitutions result mostly from head-on collisions and frequently occur at a nucleotide that is conserved in promoters recognized by the major σ factor in bacteria. This substitution is generated via adenine deamination on the template strand in the promoter open complex, as a consequence of head-on replication perturbing transcription initiation. We conclude that replication–transcription collisions induce distinct mutation signatures by antagonizing replication and transcription, not only in coding sequences but also in gene regulatory elements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
French, S. Consequences of replication fork movement through transcription units in vivo. Science 258, 1362–1365 (1992)
Liu, B. & Alberts, B. M. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267, 1131–1137 (1995)
Vilette, D., Ehrlich, S. D. & Michel, B. Transcription-induced deletions in Escherichia coli plasmids. Mol. Microbiol. 17, 493–504 (1995)
Prado, F. & Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24, 1267–1276 (2005)
Mirkin, E. V. & Mirkin, S. M. Mechanisms of transcription-replication collisions in bacteria. Mol. Cell. Biol. 25, 888–895 (2005)
Pomerantz, R. T. & O’Donnell, M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456, 762–766 (2008)
Srivatsan, A., Tehranchi, A., MacAlpine, D. M. & Wang, J. D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 6, e1000810 (2010)
Dutta, D., Shatalin, K., Epshtein, V., Gottesman, M. E. & Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146, 533–543 (2011)
Merrikh, H., Machón, C., Grainger, W. H., Grossman, A. D. & Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 470, 554–557 (2011)
Kim, N. & Jinks-Robertson, S. Transcription as a source of genome instability. Nature Rev. Genet. 13, 204–214 (2012)
Paul, S., Million-Weaver, S., Chattopadhyay, S., Sokurenko, E. & Merrikh, H. Accelerated gene evolution through replication-transcription conflicts. Nature 495, 512–515 (2013)
Fijalkowska, I. J., Jonczyk, P., Tkaczyk, M. M., Bialoskorska, M. & Schaaper, R. M. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl Acad. Sci. USA 95, 10020–10025 (1998)
Bruand, C., Bidnenko, V. & Ehrlich, S. D. Replication mutations differentially enhance RecA-dependent and RecA-independent recombination between tandem repeats in Bacillus subtilis. Mol. Microbiol. 39, 1248–1258 (2001)
Hastings, P. J., Lupski, J. R., Rosenberg, S. M. & Ira, G. Mechanisms of change in gene copy number. Nature Rev. Genet. 10, 551–564 (2009)
Kunkel, T. A. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 74, 91–101 (2009)
Rocha, E. P. C. & Danchin, A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nature Genet. 34, 377–378 (2003)
Reijns, M. A. M. et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015)
Schroeder, J. W., Hirst, W. G., Szewczyk, G. A. & Simmons, L. A. The effect of local sequence context on mutational bias of genes encoded on the leading and lagging strands. Curr. Biol. 26, 692–697 (2016)
Million-Weaver, S. et al. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc. Natl Acad. Sci. USA 112, E1096–E1105 (2015)
Luria, S. E. & Delbrück, M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943)
Rosche, W. A. & Foster, P. L. Determining mutation rates in bacterial populations. Methods 20, 4–17 (2000)
Mirkin, E. V., Castro Roa, D., Nudler, E. & Mirkin, S. M. Transcription regulatory elements are punctuation marks for DNA replication. Proc. Natl Acad. Sci. USA 103, 7276–7281 (2006)
Vilette, D., Uzest, M., Ehrlich, S. D. & Michel, B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J. 11, 3629–3634 (1992)
Tehranchi, A. K. et al. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141, 595–605 (2010)
Feklistov, A. & Darst, S. A. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147, 1257–1269 (2011)
Yoshiyama, K., Higuchi, K., Matsumura, H. & Maki, H. Directionality of DNA replication fork movement strongly affects the generation of spontaneous mutations in Escherichia coli. J. Mol. Biol. 307, 1195–1206 (2001)
Schaaper, R. M., Danforth, B. N. & Glickman, B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J. Mol. Biol. 189, 273–284 (1986)
Lu, A. L., Clark, S. & Modrich, P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl Acad. Sci. USA 80, 4639–4643 (1983)
Zhang, Y. et al. Structural basis of transcription initiation. Science 338, 1076–1080 (2012)
Zuo, Y. & Steitz, T. A. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell 58, 534–540 (2015)
Vasantha, N. & Freese, E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J. Bacteriol. 144, 1119–1125 (1980)
Janes, B. K. & Stibitz, S. Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74, 1949–1953 (2006)
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6, 343–345 (2009)
Hall, B. M., Ma, C.-X., Liang, P. & Singh, K. K. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25, 1564–1565 (2009)
Lippert, M. J. et al. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc. Natl Acad. Sci. USA 108, 698–703 (2011)
Ter Beek, A. et al. Transcriptome analysis of sorbic acid-stressed Bacillus subtilis reveals a nutrient limitation response and indicates plasma membrane remodeling. J. Bacteriol. 190, 1751–1761 (2008)
Sierro, N., Makita, Y., de Hoon, M. & Nakai, K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36, D93–D96 (2008)
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004)
Markowitz, V. M. et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122 (2012)
Gao, F. & Zhang, C.-T. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics 23, 1866–1867 (2007)
Katoh, K. & Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26, 1899–1900 (2010)
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006)
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010)
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007)
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009)
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993)
Aamodt, R. M., Falnes, P. Ø., Johansen, R. F., Seeberg, E. & Bjørås, M. The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. J. Biol. Chem. 279, 13601–13606 (2004)
Zamenhof, S. Gene unstabilization induced by heat and by nitrous acid. J. Bacteriol. 81, 111–117 (1961)
Acknowledgements
We thank E. Robleto, R. Yasbin and L. Simmons for strains, M. Cox, R. Gourse, C. Hittinger, R. Landick, K. Wasserman, C. Gross, M. Laub, S. Rosenberg, L. Simmons and the Wang laboratory for discussions and comments on the manuscript. This work was supported by the National Institutes of Health Director’s New Innovator Award DP2OD004433 to J.D.W.
Author information
Authors and Affiliations
Contributions
J.D.W. conceptualized the study. T.S.S., B.D.W. and J.D.W. designed the experiments. T.S.S. performed thyP3 fluctuation tests and sequencing of recA, yqjH, mutSL and adeC mutants, comparative genomic analyses, nitrous acid mutagenesis, competition assays and plating efficiency of mutants. B.D.W. developed the forward mutation assay, fluctuation tests and sequencing of wild-type strains, qRT–PCR, nalidixic acid fluctuation test, doubling time measurements and developed the restriction digest screening. Y.D. assisted the competition assay, plating efficiency and mutSL fluctuation tests. S.A.L. performed thyP3 fluctuation tests with B.D.W. T.S.S., B.D.W. and J.D.W. analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks S. Mirkin, E. Nudler and R. Pomerantz for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Development of a forward mutation assay that detects loss-of-function mutations in B. subtilis.
a, Simplified diagram of thymidine monophosphate (dTMP) synthesis. The phage-encoded thyP3 encodes thymidylate synthetase, which synthesizes dTMP and dihydrofolate (DHF) from dUMP and tetrahydrofolate (THF). DHF is recycled back to THF by dihydrofolate reductase (DHFR). Trimethoprim inhibits DHFR, thus blocking recycling of the essential cofactor THF and available THF is depleted by active thymidylate synthetase and cell growth is inhibited. Because cells with active thymidylate synthetase rely solely on endogenous dTMP synthesis, thyP3+ cells are sensitive to trimethoprim and loss-of-function mutations in thyP3 lead to trimethoprim resistance, which is the basis for the forward mutation assay. Viabilities of wild-type (thyA+ thyBts), thyP3+ (∆thyA thyBts thyP3+) and thyP3− (∆thyA thyBts thyP3−) cells are shown in the table and representative colonies (at 45 °C) are shown on the right. b, Competition between strains carrying wild-type (WT) and mutant thyP3 in a ∆thyA thyBts background to determine if there is any selective pressure on different mutants during growth phase at permissive temperature (37 °C). Relative fitness (mean ± s.d.) of six replicates is shown. c, Shifting the temperature to 45 °C does not affect plating efficiency during selection for trimethoprim resistance. Wild-type and mutant thyP3 cells were grown at 37 °C and plated on solid medium supplemented with IPTG plus thymine at 37 °C and 45 °C, and colony-forming units (c.f.u.) ml−1 optical density (OD600 nm)−1 was determined. Mean ± s.d. of three replicates is shown. d, thyP3 mutants have growth defects without thyBts. The doubling times of thyP3 mutant (a deletion and a frameshift mutant) in the ∆thyA ∆thyB background at 37 °C are longer, indicative of growth defects in the absence of the backup gene thyB. Mean ± s.d. of three replicates is shown. For b and d, the mutant strains are listed in Extended Data Table 1.
Extended Data Figure 2 Expression level and mutation rate of thyP3.
a, thyP3 expression in co-directional and head-on orientations. Using real-time quantitative PCR, messenger RNA level of thyP3 in the co-directional and head-on strains under induced (+IPTG) condition was measured and normalized to the reference gene accA. Since level of expression is similar between the strains, the observed difference in thyP3 mutation rate between co-directional and head-on orientations (Fig. 1d) is not caused by intrinsic differences in the expression level of thyP3. b, The orientation-specific difference in thyP3 mutation rate is not due to global increase of mutagenesis in the head-on strain. As a control to show that the increase in mutation rate is local to the thyP3 reporter, we measured the mutation rate for resistance to nalidixic acid (NalR, conferred by mutations in the gyrA gene) in co-directional and head-on strains. Since the NalR mutation rates in the two strains were similar, we conclude that the observed increase in head-on mutation rate is specific to thyP3 gene. c, Schematics of the co-directional and head-on thyP3 constructs (left) and an additional control to examine the effect of the genomic context on thyP3 mutagenesis, the neighbouring genes were swapped (right). In each construct, the thyP3 gene is flanked by the lacI gene and the spectinomycin-resistance gene. The reporter constructs were integrated into the chromosome at the amyE locus by double crossover. The direction of replication is shown at the top. The co-directional-swapped strain was created by inverting the lacI-thyP3-spc from the head-on strain and the head-on-swapped construct was created by inverting the same construct from the co-directional strain. Thus the swapped constructs switch the neighbouring transcription units. The dotted lines in each construct show the swapping boundary. d, The mutation rate of the swapped head-on strain is still higher than the swapped co-directional strain when transcription is induced (+IPTG), indicating that the difference in mutation rate between reporter strains is not due to the direction of thyP3 relative to its neighbouring genes. e, Mutation rate of co-directional and head-on thyP3 under uninduced (−IPTG) and induced (+IPTG) transcription. The rate of each class of mutations obtained under each condition is also depicted within each bar. For b, d and e, mean ± s.e.m. of n ≥ 3 independent experiments is shown. **P < 0.01, Student’s t-test.
Extended Data Figure 3 Mutation spectra of thyP3 under induced transcription.
a, b, Illustrations of the mutation spectra of the thyP3 mutants obtained from fluctuation tests of co-directional (n = 214) (a) and head-on (n = 232) (b) strains when transcription is induced (+IPTG). The thyP3 coding sequence with its promoter is shown. Sequence coordinates are indicated with reference to +1 transcription start site. The symbols used to represent different mutations are shown at the bottom, and base substitutions are shown in blue above the sequence. The numbers marked in orange next to a mutation denote the frequency. The promoter elements, Shine–Dalgarno (SD) sequence, and start and stop codons are highlighted in each spectrum.
Extended Data Figure 4 Mutation spectra of thyP3 under uninduced transcription.
a, b, Illustrations of the mutation spectra of the thyP3 mutants obtained from fluctuation tests of co-directional (n = 163) (a) and head-on (n = 178) (b) strains when transcription is not induced (−IPTG). The thyP3 coding sequence with its promoter is shown. Sequence coordinates are indicated with reference to +1 transcription start site. The symbols used to represent different mutations are shown at the bottom, and base substitutions are shown in blue above the sequence. The numbers marked in orange next to a mutation denote the frequency. The promoter elements, Shine–Dalgarno (SD) sequence, and start and stop codons are highlighted in each spectrum.
Extended Data Figure 5 Absence of selection bias in thyP3 forward mutation assay.
a–c, Growth competition experiments were performed between the C−7 promoter mutant against the following mutants: missense mutant (a), nonsense mutant (b) and frameshift mutant (c). Each mutant was competed against the C−7 promoter mutant to check if there is a competitive disadvantage for a mutant that has a mutation within the coding sequence, which may explain the high frequency of C−7 mutation compared to other mutations. The results show no fitness disadvantage for any of the mutants tested, suggesting that the high frequency of C−7 promoter mutation is not due to a selection bias. Mean ± s.d. of six replicates is shown and mutants competed are indicated within the plot. d, Plating efficiency of different thyP3 mutants. Plating efficiency was determined to check whether different classes of thyP3 mutants have differences in their plating efficiency on trimethoprim-selection plates at 45 °C, which may explain the variation in the mutation rates and spectrum. The result shows similar plating efficiency among the different mutants, suggesting that plating efficiency does not underlie the variation in the observed mutation rates. The different mutants tested are indicated on the x axis. Mean ± s.d. of three replicates is shown. The mutant strains are listed in the Extended Data Table 1.
Extended Data Figure 6 Mechanism of indel generation.
a, Representative deletion and duplication events in thyP3. A high-frequency deletion and duplication event observed in the thyP3 gene in co-directional and head-on strains. The sequence coordinates are denoted and repeat sequence is underlined. b, Table showing the mutation rate of indels (≥3 bp) in intragenic region and promoter normalized by the length of the region suggests that the localized rate of indels is higher in the promoter than the intragenic region. c, First encounter between replication and transcription machineries generates indels. Model describing the first-encounter hypothesis proposed on the basis of results presented in Fig. 2a–f. In co-directional orientation under induced transcription (+IPTG), when an array of RNA polymerases (RNAPs) transcribe the gene, the replisome is likely to collide with the first transcription complex at the promoter or promoter-proximal regions. By contrast, when transcription is induced in head-on orientation, the replisome encounters the first transcription complex from the 3′ end. In support of this first-encounter model, when transcription is not induced (basal level) the density of RNAP is sparse along the gene, hence the sites of collisions are altered. In addition, it is possible that under basal transcription, replisome collides with either the RNAP complex arrested at the promoter or with the Lac repressor, which may explain the relatively high frequency of deletion at the promoter. Thus the first-encounter model of replication–transcription collisions supports the idea that collisions stall replisome progression, triggering indel mutations. d, Mutation rate of insertions and deletions (≥3 bp) within 5′ or 3′ half of the intragenic region. Mean ± s.e.m. of n ≥ 3 experiments is shown. e, Models illustrating the different pathways that can lead to generation of indels after head-on collision-induced replication stalling: slippage, fork-reversal or template switching. f, Illustration of a complex mutation observed in thyP3 that is probably generated via microhomology-mediated break-induced replication. The complex mutation encompassing a deletion and insertion of an inverted region was observed under induced transcription in head-on orientation. The sequence coordinates are marked on the top with reference to the transcription start site (+1).
Extended Data Figure 7 Role of recombination protein RecA in collision-induced mutations.
a, Mutation rates of co-directional and head-on thyP3 strains for trimethoprim resistance in ∆recA background. Similar to wild type the mutation rate of head-on is higher than the co-directional strain, although the total rate of mutation is decreased in a ∆recA background. b, Rate of ≥3 bp indels at the promoter in co-directional orientation is strongly decreased in ∆recA cells, suggesting that indels at the promoter are mostly RecA-dependent. c, Intragenic distribution of ≥3 bp indels in ∆recA is similar to the distribution observed for wild type (Fig. 2f), thus suggesting that RecA is not necessary for the collision-induced indels within the coding region. d, Mutation rate of base substitutions in ∆recA cells is higher in head-on than co-directional orientation. e, The rate of T−7→C−7 mutation is higher in head-on relative to co-directional orientation in ∆recA cells, thus promoter substitutions can occur at a higher rate independent of recombination-mediated repair. All the fluctuation tests in a ∆recA background were performed under inducing conditions (+IPTG). Mean ± s.e.m. of n ≥ 3 experiments is shown. **P < 0.01; ***P < 0.001; Student’s t-test.
Extended Data Figure 8 Base substitutions and the role of mismatch repair and enzymatic adenine deamination.
a, IPTG induction does not affect the base substitution rate in the coding region of thyP3 when considering identical target sites, indicating that collisions may not be a major source of these mutations. In yeast, it was shown that transcription-associated mutagenesis is proportional to the level of transcription10. In B. subtilis, the total rate of base substitutions in the coding region decreases upon IPTG induction, which could be due to an unidentified transcription-dependent mutation-correction mechanism, or due to increase of target size of base substitutions in the coding sequence in uninduced (basal) transcription. b, Table showing the rates of base substitutions in the coding region and promoter of thyP3 normalized by length of the region. Localized substitution rates are higher in the promoter than coding sequence, thus suggesting that collision has a more drastic effect on promoter substitutions. c, Comparative genomic analysis of mutation rates of promoters with and without repressor binding. Nucleotide diversity per site (theta) was calculated for each promoter across different strains of B. subtilis. The comparison shows no significant difference in nucleotide diversity between repressor-bound promoters and the rest of the promoters, indicating that repressor binding may not affect the substitution rate of a promoter. Whole genomes and the repressors analysed are listed in Extended Data Table 2. NS, not significant, P > 0.05; Mann–Whitney U-test. d, The mutation frequency of T−7→C−7 mutation is higher in head-on than co-directional orientation in E. coli. The mutation frequency was calculated here from the plasmid-based forward mutation assay data reported previously26. e, The restriction-digestion-based assay to screen for T−7→C−7 mutation. Wild-type promoter sequence does not have an AflIII restriction site, whereas the promoter T−7→C−7 mutation will be digested by AflIII, which is illustrated by a representative agarose gel. f, Mismatch repair mutant (mutSL::kan) shows an expected increase (~60-fold) in total mutation rate of thyP3 in both co-directional and head-on orientation compared to wild-type. The mutation rates of the wild-type strains are presented in Fig. 1d. g, Mismatch repair mutant shows a marked ~1,000-fold increase in mutation rate of T→C substitution hotspots within the coding sequence of head-on thyP3, indicating that mismatch repair corrects T→C substitution within the coding sequence. h, Deletion of adeC gene encoding adenine deaminase modestly reduces the mutation rate of T−7→C−7 substitution in both co-directional and head-on orientation compared with wild type. For f–h, mean ± s.e.m. of n ≥ 3 experiments is shown. *P < 0.05; ***P < 0.001; Student’s t-test.
Rights and permissions
About this article
Cite this article
Sankar, T., Wastuwidyaningtyas, B., Dong, Y. et al. The nature of mutations induced by replication–transcription collisions. Nature 535, 178–181 (2016). https://doi.org/10.1038/nature18316
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18316
This article is cited by
-
Dynamic alternative DNA structures in biology and disease
Nature Reviews Genetics (2023)
-
Effect of long-term exposure of mixture of ZnO and CuO nanoparticles on Scenedesmus obliquus
Ecotoxicology (2023)
-
Testing the adaptive hypothesis of lagging-strand encoding in bacterial genomes
Nature Communications (2022)
-
Reply to: Testing the adaptive hypothesis of lagging-strand encoding in bacterial genomes
Nature Communications (2022)
-
Genomic landscape of hepatocarcinogenesis
Journal of Human Genetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.