Abstract
Mitochondrial DNA (mtDNA) mutations are maternally inherited and are associated with a broad range of debilitating and fatal diseases1. Reproductive technologies designed to uncouple the inheritance of mtDNA from nuclear DNA may enable affected women to have a genetically related child with a greatly reduced risk of mtDNA disease. Here we report the first preclinical studies on pronuclear transplantation (PNT). Surprisingly, techniques used in proof-of-concept studies involving abnormally fertilized human zygotes2 were not well tolerated by normally fertilized zygotes. We have therefore developed an alternative approach based on transplanting pronuclei shortly after completion of meiosis rather than shortly before the first mitotic division. This promotes efficient development to the blastocyst stage with no detectable effect on aneuploidy or gene expression. After optimization, mtDNA carryover was reduced to <2% in the majority (79%) of PNT blastocysts. The importance of reducing carryover to the lowest possible levels is highlighted by a progressive increase in heteroplasmy in a stem cell line derived from a PNT blastocyst with 4% mtDNA carryover. We conclude that PNT has the potential to reduce the risk of mtDNA disease, but it may not guarantee prevention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
15 June 2016
The reviewer information statement did not display correctly online when this paper was first published; this has been corrected and the statement is now available.
References
Schon, E. A., DiMauro, S. & Hirano, M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nature Rev. Genet. 13, 878–890 (2012)
Craven, L. et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 465, 82–85 (2010)
Wallace, D. C. & Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 5, a021220 (2013)
McFarland, R. et al. Multiple neonatal deaths due to a homoplasmic mitochondrial DNA mutation. Nature Genet. 30, 145–146 (2002)
Steffann, J. et al. Data from artificial models of mitochondrial DNA disorders are not always applicable to humans. Cell Reports 7, 933–934 (2014)
Richardson, J. et al. Concise reviews: assisted reproductive technologies to prevent transmission of mitochondrial DNA disease. Stem Cells 33, 639–645 (2015)
Tachibana, M. et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature 493, 627–631 (2013)
Paull, D. et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature 493, 632–637 (2013)
McGrath, J. & Solter, D. Nuclear transplantation in the mouse embryo by microsurgery and cell fusion. Science 220, 1300–1302 (1983)
Fenwick, J., Platteau, P., Murdoch, A. P. & Herbert, M. Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum. Reprod. 17, 407–412 (2002)
Schatten, H. & Sun, Q. Y. The role of centrosomes in mammalian fertilization and its significance for ICSI. Mol. Hum. Reprod. 15, 531–538 (2009)
Niakan, K. K., Han, J., Pedersen, R. A., Simon, C. & Pera, R. A. R. Human pre-implantation embryo development. Development 139, 829–841 (2012)
Forman, E. J. et al. Oocyte vitrification does not increase the risk of embryonic aneuploidy or diminish the implantation potential of blastocysts created after intracytoplasmic sperm injection: a novel, paired randomized controlled trial using DNA fingerprinting. Fertil. Steril. 98, 644–649 (2012)
Blakeley, P. et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3151–3165 (2015)
van der Maaten, L. & Hinton, G. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008)
Hellebrekers, D. M. E. I. et al. PGD and heteroplasmic mitochondrial DNA point mutations: a systematic review estimating the chance of healthy offspring. Hum. Reprod. Update 18, 341–349 (2012)
Samuels, D. C., Wonnapinij, P. & Chinnery, P. F. Preventing the transmission of pathogenic mitochondrial DNA mutations: can we achieve long-term benefits from germ-line gene transfer? Hum. Reprod. 28, 554–559 (2013)
Facucho-Oliveira, J. M. & St John, J. C. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 5, 140–158 (2009)
Burgstaller, J. P., Johnston, I. G. & Poulton, J. Mitochondrial DNA disease and developmental implications for reproductive strategies. Mol. Hum. Reprod. 21, 11–22 (2015)
Hämäläinen, R. H. et al. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc. Natl Acad. Sci. USA 110, E3622–E3630 (2013)
Lee, H. S. et al. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. Cell Reports 1, 506–515 (2012)
Treff, N. R. et al. Blastocyst preimplantation genetic diagnosis (PGD) of a mitochondrial DNA disorder. Fertil. Steril. 98, 1236–1240 (2012)
Mitalipov, S., Amato, P., Parry, S. & Falk, M. J. Limitations of preimplantation genetic diagnosis for mitochondrial DNA diseases. Cell Reports 7, 935–937 (2014)
Sallevelt, S. C. E. H. et al. Preimplantation genetic diagnosis in mitochondrial DNA disorders: challenge and success. J. Med. Genet. 50, 125–132 (2013)
Steffann, J. et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J. Med. Genet. 43, 244–247 (2006)
Herbert, M., Kalleas, D., Cooney, D., Lamb, M. & Lister, L. Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb. Perspect. Biol. 7, a017970 (2015)
Choudhary, M. et al. Egg sharing for research: a successful outcome for patients and researchers. Cell Stem Cell 10, 239–240 (2012)
HFEA Guidance on Payments for Donors.HFEA Code of Practice Section 13 http://www.hfea.gov.uk/500.html (Human Fertilisation and Embryology Authority, 2009)
Hyslop, L. et al. A novel isolator-based system promotes viability of human embryos during laboratory processing. PLoS ONE 7, e31010 (2012)
Cutting, R., Morroll, D., Roberts, S. A., Pickering, S. & Rutherford, A. Elective single embryo transfer: guidelines for practice British Fertility Society and Association of Clinical Embryologists. Hum. Fertil. (Camb.) 11, 131–146 (2008)
Stephenson, E. L., Braude, P. R. & Mason, C. International community consensus standard for reporting derivation of human embryonic stem cell lines. Regen. Med. 2, 349–362 (2007)
Wells, D. et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J. Med. Genet. 51, 553–562 (2014)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Taylor, R. W., Taylor, G. A., Durham, S. E. & Turnbull, D. M. The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res. 29, e74 (2001)
Andrews, R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genet. 23, 147 (1999)
Greaves, L. C. et al. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 10, e1004620 (2014)
Wamaitha, S. E. et al. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 29, 1239–1255 (2015)
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 (2008)
White, H. E. et al. Accurate detection and quantitation of heteroplasmic mitochondrial point mutations by pyrosequencing. Genet. Test. 9, 190–199 (2005)
Acknowledgements
We are very grateful to those who donated gametes for this research and we thank M. Nesbitt and K. Lennox for obtaining their consent. We also thank P. Chinnery and V. Floros for helpful discussion. The work was funded by the Wellcome Trust (096919) and by grants from the National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre and the Barbour Foundation. K.K.N. and co-workers are supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council (MC_UP_1202/9) and the Wellcome Trust, and by the March of Dimes Foundation (FY11-436). D.W. and co-workers are funded by the NIHR Oxford Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
M.H. and L.A.H. conceived and designed the PNT experiments. L.A.H., L.I., L.C. and L.A. performed PNT experiments and embryo manipulations. J.R., D.K. and Q.Z. performed cell counts. D.W., E.F. and S.A. performed whole-genome amplification and array-CGH. K.K.N., P.B. and N.M.E.F. performed RNA-seq experiments. L.C., H.A.T. and D.M.T. measured mtDNA carryover and performed mtDNA haplogroup analysis. N.P., K.K.N., N.M.E.F., S.E.W., Y.T. and H.O’K. derived, cultured and characterized ES cell lines. K.K.N., P.B., M.L., J.R., L.A.H., L.C., Y.T., P.B. and M.H. analysed data. A.P.M. and M.C. coordinated the oocyte donation program. M.H. wrote the manuscript with input from D.M.T., D.W., K.K.N., J.R. and M.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information: Nature thanks J. Carroll, G. Manfredi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 ltPNT and pronuclei centralization after ePNT.
a, Schematic showing timing of ltPNT and ePNT. b, Images showing the stages of the ltPNT process: left, late pronuclear zygote; middle, enucleation; right, fusion. Scale bar, 20 μm. Note large pronuclei and pipette size compared with Fig. 1b and Supplementary Videos 1 and 2. c, Images show unmanipulated and ePNT zygotes at 16–18 h after fertilization. Scale bars, 20 μm. d, Pronuclei centralization and division to the two-cell stage assessed by live cell imaging in control and ePNT zygotes (not significant). Comparisons by χ2 test.
Extended Data Figure 2 Blastocyst morphology and effect of PNT on blastocyst development and quality.
a, Schematic showing cell lineages in a mammalian blastocyst: trophectoderm; primitive endoderm and epiblast. b, Morphological criteria and scoring system used for grading human blastocysts31. Top, degree of expansion ranging from an early, unexpanded blastocyst (score 1) to fully expanded (score 6). Middle, range of ICM morphologies from absent (score 1) to large but tightly packed (score 5). Bottom, range of trophectoderm morphologies from scant and discontinuous (score 1) to a fully formed layer of tightly packed cells (score 3). Box colours correspond to the grades shown in c. c, Table used to assign blastocyst grades, according to levels of expansion, and morphology of the ICM and trophectoderm. Grade F (not shown) was assigned to embryos that developed to the blastocyst stage but subsequently showed signs of degeneration. d, Graph showing the relationship between blastocyst grade and implantation. Data obtained from clinical IVF cycles (n = 531) in which unmanipulated single blastocysts were replaced on day 5. Implantation is defined by the detection of a fetal heartbeat at 6 weeks after IVF treatment. There was no case in which a grade D or F blastocyst was replaced. P values are shown (χ2 test). e, ltPNT experimental conditions, blastocyst formation (P < 0.01; χ2 test) and quality. A total of 51 zygotes from 10 donors were allocated either to an unmanipulated control group (Ctr.; n = 12) or to ltPNT involving transfer between pairs of zygotes from the same donor (n = 29) or replacement back into the same zygote (autologous PNT (Atlg.) n = 10). f, ePNT (series I) experimental conditions, blastocyst formation and quality. This series of experiments involved a total of 58 zygotes from 13 donors. Zygotes were allocated to a control group (n = 19), or to ePNT involving either autologous (n = 18) or heterologous (Het.; n = 21) transfers. Differences are not significant (χ2 test and Fisher’s exact test). g, Image of an ePNT blastocyst fixed on day 6 and stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 50 μm. h, Cell number assessed by nuclear counts showing comparable numbers in control and ePNT (series II) blastocysts and significantly reduced numbers in ltPNT and ePNT (series I) blastocysts (P = 0.001; one-way analysis of variance (ANOVA) with Tukey’s HSD test). Mean ± standard deviation (s.d.) calculated from individual blastocysts, numbers indicated on the x-axis.
Extended Data Figure 3 Survival and blastocyst development after ePNT between zygotes obtained from freshly harvested and vitrified oocytes.
a, Experimental scheme for heterologous ePNT in series II. Because of unpredictability in the response to ovarian stimulation, heterologous transfers involved reciprocal ePNT between zygotes generated from freshly harvested and vitrified oocytes. This resulted in reconstituted zygotes whose cytoplasts originated from a fresh oocyte (FreshCy), or from a vitrified oocyte (VitCy). Oocytes for these experiments were vitrified predominantly at the MII stage (blue box; n = 80 zygotes; 25 donors). We also conducted a series of experiments to determine whether vitrification at the 2PB stage (green box; n = 34 zygotes; 13 donors) would promote improved blastocyst formation. b, Survival of reconstituted zygotes as a proportion of those submitted to autologous (Atlg.) and heterologous (Het.) ePNT according to the stage of vitrification (MII or 2PB) and according to whether the cytoplast was derived from a fresh (FreshCy) or a vitrified (VitCy) oocyte. Loss was generally due to karyoplast lysis, excessive leakage of cytoplasm, or degeneration of reconstituted zygotes during subsequent incubation. Differences are not significant (χ2 test). c, Sucrose was initially included in the manipulation medium to facilitate enucleation and fusion, however, it was later removed because the data indicated that the osmotic effect resulted in increased mtDNA carryover (see Fig. 4b). Omission of sucrose from the enucleation medium had a small, but not significant, effect on survival of zygotes whose cytoplasts originated from vitrified MII oocytes (χ2 test). d, Blastocyst formation as a percentage of zygotes submitted to the ePNT procedure recorded on days 5 and 6 after fertilization. e, Blastocyst formation recorded on days 5 and 6 as a percentage of zygotes that survived the ePNT procedure. The numbers in each group and P values are shown, χ2 test. f, Blastocyst quality grades (see Extended Data Fig. 2c, d) on days 5 and 6 (not significant; Fisher’s exact test). Source data files are available online.
Extended Data Figure 4 Array-CGH results for PNT blastocysts.
Summary of array-CGH results obtained from ICM and trophectoderm samples from control (n = 11) and ePNT (n = 30) blastocysts. Blastocysts are ordered by grade within experimental groups. The karyoplast donor age is also shown.
Extended Data Figure 5 Experimental approach and bioinformatics analysis of single-cell RNA-seq data from ePNT and control blastocysts.
a, Flow diagram showing the steps involved in RNA-seq of single cells microdissected from human blastocysts. b, Summary table of control and ePNT blastocysts submitted for RNA-seq analysis. For the purpose of gene expression analysis, we distinguish between ePNT blastocysts derived from fusion of cytoplasts and karyoplasts with the same, and different, mitochondrial genomes. Those with the same mitochondrial genomes included blastocysts from autologous ePNT and from a zygote pair donated by two sisters, which we refer to as homologous ePNT. Blastocysts arising from heterologous ePNT represent new combinations of nuclear and mitochondrial genome and are subgrouped according to the cytoplast origin (see Extended Data Fig. 4). c, Flow diagram outlining the bioinformatics analysis of RNA-seq data. Data were normalized either as reads per kilobase per million mapped reads (RPKM)41 or using DESeq2 (ref. 36). Normalized data were used to generate PCA plots, t-SNE plots and heatmaps.
Extended Data Figure 6 Analysis of differential gene expression in good quality ePNT and control blastocysts.
a, PCA matrix using the first ten principal components of DESeq2 VST normalized data for the top 12,000 most variable genes. Global gene expression is indistinguishable between unmanipulated control and ePNT samples, PC1 versus PC2 highlighted in blue box. b, PCA matrix as shown in a, distinguished by lineage, clearly seen in PC2 versus PC3 (pink box). c, t-SNE analysis after DESeq2 VST normalization of 6,000 of the most variably expressed genes, where samples were distinguished by lineage. Sample numbers and blastocyst grades are shown. Autologous and homologous ePNT samples are derived from blastocysts in which the karyoplast and cytoplast had the same mitochondrial genome. Heterologous ePNT samples were derived from pairs of zygotes with different mitochondrial genomes (Extended Data Fig. 5). Samples from experimental controls and reference population were combined for the purpose of the analyses shown in a and b.
Extended Data Figure 7 Expression of lineage-specific genes and mitochondrial OXPHOS genes in control and ePNT embryos.
a, Heatmap showing log2-transformed RPKM values of selected differentially expressed genes in trophectoderm (n = 10), epiblast (n = 10) and primitive endoderm (n = 10) lineages. b, Heatmap showing expression of mitochondrial OXPHOS genes after unsupervised hierarchical clustering. Expression of OXPHOS genes encoded by mtDNA is variable both within and between blastocysts. Control and ePNT samples cluster together, irrespective of whether the cytoplast and karyoplast had the same (blue font) or different (purple font) mtDNA. Sample numbers and blastocyst grades are shown. The reference population includes a previously published series14. Autologous and homologous ePNT samples are derived from blastocysts in which the karyoplast and cytoplast had the same mitochondrial genome. Heterologous ePNT samples were derived from pairs of zygotes with different mitochondrial genomes (Extended Data Fig. 5b). Expression levels are indicated on a high-to-low scale (purple–white–green). Source data files are available online for a and b.
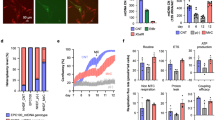
Extended Data Figure 8 Measurement of heteroplasmy due to mtDNA carryover during ePNT.
a, Mitochondrial genotypes were determined by identifying polymorphic variants in the hypervariable mtDNA control regions of each donor. Sequence electropherograms of mtDNA non-coding control region with a sequence variant used for pyrosequencing (highlighted) (m.73A>G). b, Sequence pyrograms for the mtDNA variant (m.73A>G) in control samples. The expected level of variant is given along with the level determined by pyrosequencing (in brackets). c, Examples of the standard curve generated to increase accuracy in detecting low levels (0–25%) of heteroplasmy by pyrosequencing, which has previously been reported to accurately detect heteroplasmy at a level of 1% (ref. 42). Each data point represents the mean of 3–4 technical replicates. d, mtDNA carryover was measured by pyrosequencing using clumps of cells (n = 92) from day 6 blastocysts (n = 40; names shown on y axis). The cells were predominantly obtained from the trophectoderm (TE) layer (purple, n = 67). ICM cells (red, n = 5) and cells of mixed trophectoderm/ICM origin (green, n = 20) were also analysed. Each data point represents the mean of 2–3 technical replicates. e, mtDNA carryover from individual blastocysts calculated from data shown in d. Each data point represents either the mean value where more than one sample was tested (n = 28 ePNT blastocysts), or a single value where only one sample was tested (n = 12 ePNT blastocysts). Horizontal lines show median values for each experimental group. Blastocysts arising from ePNT performed in the absence of sucrose and fused with a fresh cytoplast (FreshCy) had significantly reduced mtDNA carryover compared with blastocysts where ePNT was performed in the presence of sucrose (P values and blastocyst numbers are shown; two-sided Mann–Whitney U-test). f, Graph showing the proportions of blastocysts (n = 40) with mtDNA carryover measurements falling within the specified levels (not significant: χ2 test). Source data files are provided for c–f.
Extended Data Figure 9 Derivation and characterization of human ES cells from control and ePNT blastocysts.
a, Examples of outgrowths formed following explantation of the ICM from ePNT (n = 15) and control (n = 6) blastocysts used for hES cell derivations. The dashed white circle indicates the region picked for initial passage of the ICM outgrowth. Bottom, examples of hES cell colonies. b, Example of a normal karyogram from an PNT-hES cell line (45PNT); 4/4 lines tested showed a normal karyotype. The remaining hES cell line did not grow beyond passage 2 and was derived from a uniformly aneuploid blastocyst (55PNT; Extended Data Fig. 4). c, Immunostaining of representative PNT-hES cells (grown in mTeSR1) for NANOG, SSEA4 (green), SOX2 and OCT4 (red) with DAPI (blue) merge. Graph shows quantitative polymerase chain reaction with reverse transcription (qRT–PCR) analysis of control and PNT-hES cell lines for pluripotency transcripts NANOG, POU5F1 and SOX2. Horizontal line shows the median value, which is similar between hES cells from unmanipulated control blastocysts (Ctr.; n = 2 hES cell lines) and ePNT-hES cells (n = 4 ePNT-hES cell lines). d, Immunostaining of representative PNT-hES cells after 20 days in basal MEF media, confirming differentiation into all three germ layers: endoderm (α-fetoprotein (AFP); SOX17), mesoderm (α-smooth muscle actin (SMA); desmin (DES)) and ectoderm (β-III tubulin (TUJ1); SOX1) in green or red, with DAPI (blue) merge. Scale bars, 50 μm. e, Table showing the mtDNA variants and primers used to measure mtDNA carryover in PNT-hES cell lines. f, Summary table showing details of blastocysts and the corresponding hES cells. Aneuploidy in PNT-hES cell lines was analysed by metaphase spreads, except for 31PNT-hES, which was determined by array-CGH.
Extended Data Figure 10 Heteroplasmy in subclones of the hES cell line derived from 36PNT.
a, The 36PNT hES cell line was frozen at passage 3 (after derivation), thawed and subcloned to monitor heteroplasmy arising from the karyoplast donor mtDNA. Six colonies (15–20) were randomly selected at the first post-thaw passage (P3) and clumps of cells were plated on 3 tissue culture wells; 5/6 colonies gave rise to 3 subclones, which were grown to P11. Subclones are distinguished by colour in the graphs. Each data point represents the mean of two technical replicates for a single cell sample. Source data file is available online.
Supplementary information
The ePNT procedure in human zygotes
Video showing pronuclei being extracted in separate karyoplasts from a human zygote before being placed underneath the zona pellucida of a previously enucleated zygote. (MP4 23594 kb)
Removal of excess cytoplasm from the karyoplast during ePNT in human zygotes
Video showing karyoplast removal and shearing of excess cytoplasm in preparation for fusion with a previously enucleated zygote. (MP4 27801 kb)
Rights and permissions
About this article
Cite this article
Hyslop, L., Blakeley, P., Craven, L. et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature 534, 383–386 (2016). https://doi.org/10.1038/nature18303
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18303
This article is cited by
-
Secondary structure of the human mitochondrial genome affects formation of deletions
BMC Biology (2023)
-
Mitochondrial DNA editing with mitoARCUS
Nature Metabolism (2023)
-
Measurements of Electrical Characteristics of Mammalian Oocytes
Measurement Techniques (2023)
-
Mitochondrial adaptation in cancer drug resistance: prevalence, mechanisms, and management
Journal of Hematology & Oncology (2022)
-
Mitochondria: emerging therapeutic strategies for oocyte rescue
Reproductive Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.