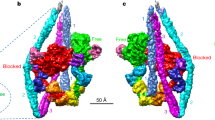
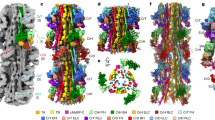
Abstract
The interaction of myosin with actin filaments is the central feature of muscle contraction1 and cargo movement along actin filaments of the cytoskeleton2. The energy for these movements is generated during a complex mechanochemical reaction cycle3,4. Crystal structures of myosin in different states have provided important structural insights into the myosin motor cycle when myosin is detached from F-actin5,6,7. The difficulty of obtaining diffracting crystals, however, has prevented structure determination by crystallography of actomyosin complexes. Thus, although structural models exist of F-actin in complex with various myosins8,9,10,11, a high-resolution structure of the F-actin–myosin complex is missing. Here, using electron cryomicroscopy, we present the structure of a human rigor actomyosin complex at an average resolution of 3.9 Å. The structure reveals details of the actomyosin interface, which is mainly stabilized by hydrophobic interactions. The negatively charged amino (N) terminus of actin interacts with a conserved basic motif in loop 2 of myosin, promoting cleft closure in myosin. Surprisingly, the overall structure of myosin is similar to rigor-like myosin structures in the absence of F-actin, indicating that F-actin binding induces only minimal conformational changes in myosin. A comparison with pre-powerstroke and intermediate (Pi-release)7 states of myosin allows us to discuss the general mechanism of myosin binding to F-actin. Our results serve as a strong foundation for the molecular understanding of cytoskeletal diseases, such as autosomal dominant hearing loss and diseases affecting skeletal and cardiac muscles, in particular nemaline myopathy and hypertrophic cardiomyopathy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The coordinates and electron microscopy density maps have been deposited in the Protein Data Bank (PDB) under accession numbers 5JLF and 5JLH and the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-8162 to EMD-8165.
References
Huxley, H. E. The mechanism of muscular contraction. Science 164, 1356–1366 (1969)
Heissler, S. M. & Manstein, D. J. Nonmuscle myosin-2: mix and match. Cell. Mol. Life Sci. 70, 1–21 (2013)
Lymn, R. W. & Taylor, E. W. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry 10, 4617–4624 (1971)
Sweeney, H. L. & Houdusse, A. Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 39, 539–557 (2010)
Coureux, P.-D., Sweeney, H. L. & Houdusse, A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 23, 4527–4537 (2004)
Gulick, A. M., Bauer, C. B., Thoden, J. B. & Rayment, I. X-ray structures of the MgADP, MgATPγS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry 36, 11619–11628 (1997)
Llinas, P. et al. How actin initiates the motor activity of myosin. Dev. Cell 33, 401–412 (2015)
Lorenz, M. & Holmes, K. C. The actin-myosin interface. Proc. Natl Acad. Sci. USA 107, 12529–12534 (2010)
Rayment, I. et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science 261, 58–65 (1993)
Behrmann, E. et al. Structure of the rigor actin-tropomyosin-myosin complex. Cell 150, 327–338 (2012)
Holmes, K. C., Angert, I., Kull, F. J., Jahn, W. & Schröder, R. R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425, 423–427 (2003)
von der Ecken, J. et al. Structure of the F-actin–tropomyosin complex. Nature 519, 114–117 (2015)
Várkuti, B. H. et al. A novel actin binding site of myosin required for effective muscle contraction. Nature Struct. Mol. Biol. 19, 299–306 (2012)
Onishi, H., Mikhailenko, S. V. & Morales, M. F. Toward understanding actin activation of myosin ATPase: the role of myosin surface loops. Proc. Natl Acad. Sci. USA 103, 6136–6141 (2006)
Furch, M., Remmel, B., Geeves, M. A. & Manstein, D. J. Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry 39, 11602–11608 (2000)
Sasaki, N., Asukagawa, H., Yasuda, R., Hiratsuka, T. & Sutoh, K. Deletion of the myopathy loop of Dictyostelium myosin II and its impact on motor functions. J. Biol. Chem. 274, 37840–37844 (1999)
Joel, P. B., Trybus, K. M. & Sweeney, H. L. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J. Biol. Chem. 276, 2998–3003 (2001)
Murphy, C. T. & Spudich, J. A. The sequence of the myosin 50–20K loop affects myosin’s affinity for actin throughout the actin–myosin ATPase cycle and its maximum ATPase activity. Biochemistry 38, 3785–3792 (1999)
Uyeda, T. Q. P., Ruppel, K. M. & Spudich, J. A. Enzymatic activities correlate with chimaeric substitutions at the actin-binding face of myosin. Nature 368, 567–569 (1994)
Kojima, S. et al. Functional roles of ionic and hydrophobic surface loops in smooth muscle myosin: their interactions with actin. Biochemistry 40, 657–664 (2001)
Milligan, R. A., Whittaker, M. & Safer, D. Molecular structure of F-actin and location of surface binding sites. Nature 348, 217–221 (1990)
Van Dijk, J., Furch, M., Lafont, C., Manstein, D. J. & Chaussepied, P. Functional characterization of the secondary actin binding site of myosin II. Biochemistry 38, 15078–15085 (1999)
Várkuti, B. H., Yang, Z. & Malnasi-Csizmadia, A. Structural model of weak binding actomyosin in the prepowerstroke state. J. Biol. Chem. 290, 1679–1688 (2015)
Cook, R. K., Root, D., Miller, C., Reisler, E. & Rubenstein, P. A. Enhanced stimulation of myosin subfragment 1 ATPase activity by addition of negatively charged residues to the yeast actin NH2 terminus. J. Biol. Chem. 268, 2410–2415 (1993)
Miller, C. J., Wong, W. W., Bobkova, E., Rubenstein, P. A. & Reisler, E. Mutational analysis of the role of the N terminus of actin in actomyosin interactions. Comparison with other mutant actins and implications for the cross-bridge cycle. Biochemistry 35, 16557–16565 (1996)
Sutoh, K., Ando, M., Sutoh, K. & Toyoshima, Y. Y. Site-directed mutations of Dictyostelium actin: disruption of a negative charge cluster at the N terminus. Proc. Natl Acad. Sci. USA 88, 7711–7714 (1991)
Coureux, P.-D. et al. A structural state of the myosin V motor without bound nucleotide. Nature 425, 419–423 (2003)
Klein, J. C. et al. Actin-binding cleft closure in myosin II probed by site-directed spin labeling and pulsed EPR. Proc. Natl Acad. Sci. USA 105, 12867–12872 (2008)
Sasaki, N., Ohkura, R. & Sutoh, K. Insertion or deletion of a single residue in the strut sequence of Dictyostelium myosin II abolishes strong binding to actin. J. Biol. Chem. 275, 38705–38709 (2000)
Muretta, J. M., Rohde, J. A., Johnsrud, D. O., Cornea, S. & Thomas, D. D. Direct real-time detection of the structural and biochemical events in the myosin power stroke. Proc. Natl Acad. Sci. USA 112, 14272–14277 (2015)
Müller, M. et al. Functional characterization of the human α-cardiac actin mutations Y166C and M305L involved in hypertrophic cardiomyopathy. Cell. Mol. Life Sci. 69, 3457–3479 (2012)
Ohki, T., Ohno, C., Oyama, K., Mikhailenko, S. V. & Ishiwata, S. Purification of cytoplasmic actin by affinity chromatography using the C-terminal half of gelsolin. Biochem. Biophys. Res. Commun. 383, 146–150 (2009)
Coulton, A., Lehrer, S. S. & Geeves, M. A. Functional homodimers and heterodimers of recombinant smooth muscle tropomyosin. Biochemistry 45, 12853–12858 (2006)
Heissler, S. M. & Manstein, D. J. Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J. Biol. Chem. 286, 21191–21202 (2011)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Hohn, M. et al. SPARX, a new environment for cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007)
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Scheres, S. H. W. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3, e03665 (2014)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010)
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nature Methods 9, 853–854 (2012)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Münnich, S., Pathan-Chhatbar, S. & Manstein, D. J. Crystal structure of the rigor-like human non-muscle myosin-2 motor domain. FEBS Lett. 588, 4754–4760 (2014)
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Lopéz-Blanco, J. R. & Chacón, P. iMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J. Struct. Biol. 184, 261–270 (2013)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015)
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D 70, 2487–2499 (2014)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Anandakrishnan, R., Aguilar, B. & Onufriev, A. V. H. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541 (2012)
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005)
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
Stenson, P. D. et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133, 1–9 (2014)
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015)
Dausse, E. et al. Familial hypertrophic cardiomyopathy. Microsatellite haplotyping and identification of a hot spot for mutations in the beta-myosin heavy chain gene. J. Clin. Invest. 92, 2807–2813 (1993)
Richard, P. et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 107, 2227–2232 (2003)
Yu, B. et al. Denaturing high performance liquid chromatography: high throughput mutation screening in familial hypertrophic cardiomyopathy and SNP genotyping in motor neurone disease. J. Clin. Pathol. 58, 479–485 (2005)
Epstein, N. D., Cohn, G. M., Cyran, F. & Fananapazir, L. Differences in clinical expression of hypertrophic cardiomyopathy associated with two distinct mutations in the β-myosin heavy chain gene. A 908Leu→Val mutation and a 403Arg→Gln mutation. Circulation 86, 345–352 (1992)
Geisterfer-Lowrance, A. A. et al. A molecular basis for familial hypertrophic cardiomyopathy: a β cardiac myosin heavy chain gene missense mutation. Cell 62, 999–1006 (1990)
Blanchard, E., Seidman, C., Seidman, J. G., LeWinter, M. & Maughan, D. Altered crossbridge kinetics in the αMHC403/+ mouse model of familial hypertrophic cardiomyopathy. Circ. Res. 84, 475–483 (1999)
Cuda, G., Fananapazir, L., Zhu, W. S., Sellers, J. R. & Epstein, N. D. Skeletal muscle expression and abnormal function of beta-myosin in hypertrophic cardiomyopathy. J. Clin. Invest. 91, 2861–2865 (1993)
Woo, A. et al. Mutations of the beta-myosin heavy chain gene in hypertrophic cardiomyopathy: critical functional sites determine prognosis. Heart 89, 1179–1185 (2003)
Van Driest, S. L. et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 44, 602–610 (2004)
Mohiddin, S. A. et al. Utility of genetic screening in hypertrophic cardiomyopathy: prevalence and significance of novel and double (homozygous and heterozygous) β-myosin mutations. Genet. Test. 7, 21–27 (2003)
Marian, A. J. et al. A transgenic rabbit model for human hypertrophic cardiomyopathy. J. Clin. Invest. 104, 1683–1692 (1999)
Lankford, E. B., Epstein, N. D., Fananapazir, L. & Sweeney, H. L. Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J. Clin. Invest. 95, 1409–1414 (1995)
Perrot, A. et al. Prevalence of cardiac beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J. Mol. Med. 83, 468–477 (2005)
Erdmann, J. et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin. Genet. 64, 339–349 (2003)
Moolman, J. C., Brink, P. A. & Corfield, V. A. Identification of a new missense mutation at Arg403, a CpG mutation hotspot, in exon 13 of the β-myosin heavy chain gene in hypertrophic cardiomyopathy. Hum. Mol. Genet. 2, 1731–1732 (1993)
Moolman-Smook, J. C., De Lange, W. J., Bruwer, E. C., Brink, P. A. & Corfield, V. A. The origins of hypertrophic cardiomyopathy-causing mutations in two South African subpopulations: a unique profile of both independent and founder events. Am. J. Hum. Genet. 65, 1308–1320 (1999)
Greber-Platzer, S. et al. Beta-myosin heavy chain gene mutations and hypertrophic cardiomyopathy in Austrian children. J. Mol. Cell. Cardiol. 33, 141–148 (2001)
Villard, E. et al. Mutation screening in dilated cardiomyopathy: prominent role of the beta myosin heavy chain gene. Eur. Heart J. 26, 794–803 (2005)
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008)
Toydemir, R. M. et al. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nature Genet. 38, 561–565 (2006)
Kamisago, M. et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 343, 1688–1696 (2000)
Daehmlow, S. et al. Novel mutations in sarcomeric protein genes in dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 298, 116–120 (2002)
Acknowledgements
We thank O. Hofnagel for assistance in cryo sample preparation. We acknowledge R. Matadeen and S. de Carlo for image acquisition at the Netherlands Centre for Nanoscopy in Leiden. We thank R. S. Goody for reading the manuscript. This work was supported by the Max Planck Society, the European Research Council under the European Union’s Seventh Framework Program (FP7/2007-2013) (grant number 615984) (to S.R.), the Behrens-Weise foundation (to S.R.) and German Research Foundation (DFG) grant MA 1081/21-1 (to D.J.M.). J.v.d.E. is a fellow of Studienstiftung des deutschen Volkes.
Author information
Authors and Affiliations
Contributions
D.J.M. and S.R. designed the project. S.M.H. and S.P.-C. purified actin, tropomyosin, and myosin constructs. D.J.M. supervised protein work. J.v.d.E. prepared specimens, recorded, analysed and processed the data, and prepared figures. S.R. managed the project. J.v.d.E. and S.R. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks E. Nogales, J. Löwe and A. Houdusse for their contribution to the peer review of this work.
Extended data figures and tables
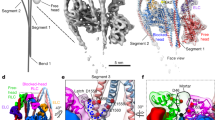
Extended Data Figure 1 Micrographs, two-dimensional classifications and three-dimensional refinement.
a–c, Representative of ~2,000 digital micrographs (a) and of 200 two-dimensional class averages of the F-actin–tropomyosin–myosin data set before (b) and after (c) three-dimensional refinement, respectively. Lower part of the micrograph is band-pass filtered to allow a better visualization of the filaments. Only filaments in rectangular boxes were chosen for refinement and bundled filaments were sorted out. d, e, Box dimension and angular distribution during three-dimensional refinement in side (d) and top (e) views. Histogram (few in blue to many in red) shows distribution of projection direction of each boxed segment relative to the three-dimensional reconstruction (grey). f, Example of two class averages out of c that show secondary structure elements. g, Fit of F-actin–myosin model to assign characteristic domains of myosin (see coloured circles and boxes). h, i, Representative of 200 class averages of the reprocessed F-actin–tropomyosin data set before (h) and after (i) three-dimensional refinement. j, k, In class averages, secondary structure elements in F-actin (green) and the coiled-coil structure of tropomyosin (yellow) are visible. Scale bars in micrograph and class averages are 50 nm and 10 nm, respectively.
Extended Data Figure 2 Resolution and model refinement of the actomyosin complex.
a, FSC curves of the cryo-EM reconstruction of the F-actin–tropomyosin–myosin data set (blue) and the reprocessed data set of F-actin–tropomyosin (red). The average resolution (FSC0.143) of the final electron density maps (central parts, green in subfigures) is estimated at 3.9 Å and 3.6 Å, respectively. Next subfigures illustrate only the actomyosin data set. b, Colour-coded local resolution of the full map and only finally refined part of the map (see Methods) estimated by ResMap43. c–e, Representative regions with higher than the average resolution in the F-actin filament core (c), the average resolution at the interface (d) and lower resolution in outer myosin parts (e). f, FSC curves of the model to each half map to check for overfitting, when the model was only refined versus the first half map. Black curve shows FSC between refined model and full map, when the model was refined against the full map (see Methods). g, h, B-factor distribution of final model from low (blue) to high (red) values. The absolute value strongly depends on the sharpening factor of the map, while the distribution shows the same gradient as the local resolution in b.
Extended Data Figure 3 HLH motif bound to F-actin.
a, Front view of F-actin and the HLH motif of the L50 domain of myosin show only small changes in loop regions while helices do not alter between weak (PPS state in purple, PDB accession number 5I4E) and strong binding (rigor state in red). The D-loop is moved towards the binding interface and is stabilized (A-state in yellow, M-state in green). Arrows indicate changes and scale bar is given. b, Same view as before shows the interface of myosin and F-actin in the rigor state. One possible salt bridge is highlighted with dotted lines. Surfaces are coloured from low (white) to high (yellow) hydrophobicity. c–e, Back view of the HLH motif and the base of loop 2 bound to central (SD1, SD3) and adjacent (D-loop) F-actin subunits. Comparison of rigor (red) and PPS state (purple, PDB accession number 5I4E) shows main differences (c). Final interaction of fully bound myosin is given in d, e. Possible electrostatic interactions are indicated by dotted lines. F-actin surface is depicted per subunit colour (c), by hydrophobicity (d) or electrostatic Coulomb potential (e, −10 kcal mol−1 in red to +10 kcal mol−1 in blue). In all subfigures, coloured residue labels belong to F-actin. f, Sequence alignment of myosin (H. sapiens myosin-II, -I, -III, -V, -VI) in the region of the HLH (helix-R–loop–helix-S) motif. Important functions at the F-actin–myosin interface and roles in stabilizing these regions themselves are highlighted and labelled. Residue numbering refers to our published structure belonging to the sequence of NM-2C (depicted in bold type). Tissue localization of myosin-II is written in parentheses. We refer to the different myosin isoforms according to the nomenclature for the genes encoding the respective myosin heavy chains.
Extended Data Figure 4 Cardiomyopathy loop and disease-causing mutations.
a, Conservation of the CM-loop in the human myosin-II class is visualized as a model on F-actin (cyan) from low (white) to high (purple) conservation. b, Sequence alignment of the CM-loop region of the human myosin-II class. Important functions at the F-actin–myosin interface are highlighted and labelled. Residue numbering refers to our published structure belonging to the sequence of NM-2C (depicted in bold type). Tissue localization of myosin-II is written in parentheses. We refer to the different myosin isoforms according to the nomenclature for the genes encoding the respective myosin heavy chains. c, d, Mutations in β-cardiac myosin (MYH7) can lead to cardiomyopathies. Corresponding residues in β-cardiac myosin are illustrated with our rigor state model (c) and known mutations57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74 are listed (d). e, Table of known disease-causing mutations at the actomyosin interface56,66,69,74,75,76,77,78. Numbers in parentheses give respective residue position in our published structure of NM-2C. Localization is described in parentheses.
Extended Data Figure 5 Loop 2 and loop 3 on F-actin.
a, b, Density map (grey) corresponding to the flexible part (residues 641–661) of loop 2 (red). F-actin is shown as surface model coloured from low (white) to high (yellow) hydrophobicity (a) or electrostatic Coulomb potential (b, −10 kcal mol−1 in red to +10 kcal mol−1 in blue). Residue labels belonging to F-actin are coloured as surface colours. c, Sequence alignment of myosin (H. sapiens myosin-II, -I, -III, -V, -VI) in the region of loop 2 and helix-R of the HLH region. Important functions at the F-actin–myosin interface and in stabilizing these regions are highlighted and labelled. Residue numbering refers to our published structure belonging to the sequence of NM-2C (depicted in bold). Tissue localization of myosin-II is written in parentheses. We refer to the different myosin isoforms according to the nomenclature for the genes encoding the respective myosin heavy chains. d, e, Changes between rigor (red) and PPS state (purple) in the loop 3 region relative to the rest of lower 50-kDa domain when bound to F-actin. Movements are indicated by black arrows and scale bars are given.
Extended Data Figure 6 Sequence-dependent interaction of supporting loop with the N terminus of F-actin.
a, Surface of myosin and N terminus is depicted by electrostatic Coulomb potential (−10 kcal mol−1 in red to +10 kcal mol−1 in blue). Involved charged residues are labelled. b, Position of the proline-rich loop (supporting loop) located between relay helix and helix-R slightly differs between the PPS (purple, PDB accession number 5I4E) and rigor state (red) and shows no direct interaction with the N terminus of F-actin. Regions at the surface of SD1 are pulled to the actomyosin interface indicated by an arrow and a scale bar is given (F-actin in A-state is depicted in yellow; F-actin in M-state is depicted in cyan). c, Sequence alignment of myosin (H. sapiens myosin-II, -I, -III, -V, -VI) in the region of the supporting loop. Different lengths of the loop and a possible supporting function are given in the last column. Residue numbering refers to our published structure belonging to the sequence of NM-2C (depicted in bold). Tissue localization of myosin-II is written in parentheses. We refer to the different myosin isoforms according to the nomenclature for the genes encoding the respective myosin heavy chains. d–g, Comparison of prominent properties in the supporting loop of different myosin classes (comparative models in purple) and their ability to undergo a direct interaction with the N terminus. Main differences are length of loop (numbers give absent amino acids relative to long loop) and position of the prominent positive-charged amino acid (R or K). Only an arginine or lysine sitting on the top would allow a direct interaction (e–g), while a sideward-oriented arginine (d) or a short loop (e) disables or reduces a possible interaction, respectively. In addition, respective densities (d) of the cryo-EM map are displayed.
Extended Data Figure 7 Myosin-induced conformational changes in F-actin.
a–c, Comparison of bare F-actin (A-state, yellow) with myosin-bound F-actin (M-state, cyan). Myosin is depicted in red. Either models (a) or representative parts of the electron density maps (b) illustrate conformational changes in F-actin (c). d, Sequence alignment of the N terminus region of human actin isoforms. Residue numbering refers to our published structure belonging to the sequence of non-muscular γ1-actin (ACTG1, depicted in bold type). To prevent confusion, the gene names instead of protein names are given. Localization is written in parentheses. e, N terminus and nucleotide binding region of F-actin undergo small changes (highlighted with arrows) through transmitted force of N terminus pulling. f, g, Close-ups of structural changes at the nucleotide binding site. h, Coordination of ADP and Mg2+ in the nucleotide binding cleft in M-state F-actin. i–k, Myosin binding induces a stabilization and shifting of the C terminus towards SD1 of F-actin. C373, which was used for pyrene labelling of F-actin, is part of the C-terminal region. Scale bars are given in the subfigures.
Extended Data Figure 8 Comparison of rigor and rigor-like myosin structures.
a–d, Close-ups of superimposed models from rigor-like structures (nucleotide-free myosin crystal structures in light green, PDB accession number 4PD3 (ref. 44) and 1OE9 (ref. 27)) with our rigor cryo-EM structure (red). F-actin is shown as surface model (green, cyan). Illustrated domains are labelled and coloured, while the rest of myosin is shown in grey from the rigor state model. Most regions do not show conformational differences (a, b), but the surface loops of myosin (CM-loop, loop 3 and loop 4) interacting with F-actin differ slightly in the rigor from the rigor-like structures (a, c). In contrast to the cryo-EM structure, loops at the interface (a, c) between F-actin and myosin are not always resolved in crystal structures. Major structural differences in the lever arm and converter regions are indicated by arrows and a scale bar is given (d).
Extended Data Figure 9 Different alignments of models for weak to strong binding of myosin and strut attraction to the base of loop2 promotes cleft closure.
a–c, Three possible alignments of myosin in the PPS (first column, purple, PDB accession number 5I4E), Pi-release (second column, blue) and rigor (third column, red) states are illustrated with respect to F-actin. For better visualization, differences in F-actin are not shown and F-actin is only depicted in the M-state (green, cyan). The Pi-release state represents a homology model of NM-2C based on a crystal structure of myosin in the Pi-release state7 (PBD accession number 4PFO, see Methods). All models are either aligned to the U50 domain (a) or the L50 domain (b) of the rigor state. In c, the model of the Pi-release state was first aligned to the L50 domain of the rigor state. The PPS state was then aligned to the U50 domain of model of the Pi-release state. The first row in each subfigure shows changes in the U50 domain from the top (for a better visualization L50 was deleted). The second row shows the L50 domain from the bottom (U50 is transparent). Possible clashes are indicated by a yellow star (a). d–f, Binding mechanism of the strut, connecting L50 and U50 domains, to the stabilized base of loop 2. To illustrate the conformational changes, the respective regions in the PPS state (PPS, purple, PDB accession number 5I4E) and rigor state (RS, red) of myosin have been partly overlaid. The rest of myosin is shown in grey. L50 binds to F-actin (A-state, yellow) (d, e). The base of loop 2 is stabilized by F-actin (e) and attracts the negatively charged strut with its positive patch. This promotes the binding of the strut, shifting the equilibrium to a closed conformational state of myosin (f). Flexible parts of loop 2 are indicated as dotted lines. Lower panels show surfaces of the same regions as in the upper panels coloured by electrostatic Coulomb potential. For better visualization, the upper parts of the strut were removed. Surface of F-actin is depicted in transparent grey.
Supplementary information
Cryo-EM structure of the ATM complex in detail
a, Cryo-EM reconstruction of F-actin (five central subunits in green and one subunit in cyan) decorated with tropomyosin (blue) and myosin in rigor state (central molecules highlighted in red). Close-up ends with the central part of the map. b, Representative part of the central core region of the F-actin filament shows better than average resolution. c, Interface between D-loop, SD1 of F-actin and HLH motif of myosin. d, Myosin CM-loop bound to SD1 and SD3 of F-actin. (MP4 9614 kb)
Model of the rigor ATM complex and subdomains in F-actin
F-actin (green, cyan) decorated with tropomyosin (blue) and myosin (red). Subdomain organisation of F-actin is shown. (MP4 8246 kb)
Domain organisation of myosin on F-actin
Domain organisation of the NM-2C head region (depicted in different colours and labelled as in Fig. 1b-d). Close-ups on HLH motif and surface loops (CM-loop, loop 3, loop 4) at the F-actin-myosin interface. (MP4 9341 kb)
F-actin-myosin interfaces in detail
a-c, Close-ups on the HLH motif (a), CM-loop (b) and loop 2 (c) of myosin bound to F-actin in rigor state. d, e, A positively charged basin is formed by loop 2, helix-W and the supporting loop of myosin to stabilize the pulled N-terminus conformation of F-actin. Myosin is shown as ribbon with residues (d) or as surface coloured by the electrostatic Coulomb potential ranging from -10 kcal mol-1 in red to +10 kcal mol-1 in blue. (MP4 7622 kb)
Model of weak to strong binding of myosin to F-actin
a, Myosin binds in a weak PPS state (purple, PDB: 5I4E) to F-actin. b, L50 rotates and binds to F-actin resulting in a stronger bound myosin (Pi-release state in blue). c, d, Finally, U50 rotates and binds to F-actin (c) obtaining the full interface of strongly bound myosin (rigor state in red) on F-actin (d). Models were aligned as shown in Extended Data Fig. 9f. For better visualization, differences in F-actin are not shown and F-actin is depicted only in the M-state (green, cyan). (MP4 5344 kb)
Model of myosin-binding to F-actin in detail
a, Initial binding of myosin (purple) to F-actin (yellow). b, L50 rotates and binds to F-actin. c, The base of loop 2 (red) is stabilized by hydrophobic interactions with F-actin. d, It also interacts with the negatively charged N-terminus of F-actin inducing its pulling and ordering (A-state in yellow, M-state in cyan). L50 is in its final rigor state conformation. e, Negatively charged strut gets attracted to the positively charged patch of the base of loop 2. U50 rotates and binds to F-actin resulting in the interface observed in the rigor state (red). (MP4 6017 kb)
Rights and permissions
About this article
Cite this article
Ecken, J., Heissler, S., Pathan-Chhatbar, S. et al. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 534, 724–728 (2016). https://doi.org/10.1038/nature18295
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18295
This article is cited by
-
Parallel cryo electron tomography on in situ lamellae
Nature Methods (2023)
-
Electron microscopy of cardiac 3D nanodynamics: form, function, future
Nature Reviews Cardiology (2022)
-
The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity
Nature Communications (2021)
-
Alpha and beta myosin isoforms and human atrial and ventricular contraction
Cellular and Molecular Life Sciences (2021)
-
Actin–tropomyosin distribution in non-muscle cells
Journal of Muscle Research and Cell Motility (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.