Abstract
Mitochondrial genomes (mitochondrial DNA, mtDNA) encode essential oxidative phosphorylation (OXPHOS) components. Because hundreds of mtDNAs exist per cell, a deletion in a single mtDNA has little impact. However, if the deletion genome is enriched, OXPHOS declines, resulting in cellular dysfunction. For example, Kearns–Sayre syndrome is caused by a single heteroplasmic mtDNA deletion. More broadly, mtDNA deletion accumulation has been observed in individual muscle cells1 and dopaminergic neurons2 during ageing. It is unclear how mtDNA deletions are tolerated or how they are propagated in somatic cells. One mechanism by which cells respond to OXPHOS dysfunction is by activating the mitochondrial unfolded protein response (UPRmt), a transcriptional response mediated by the transcription factor ATFS-1 that promotes the recovery and regeneration of defective mitochondria3,4. Here we investigate the role of ATFS-1 in the maintenance and propagation of a deleterious mtDNA in a heteroplasmic Caenorhabditis elegans strain that stably expresses wild-type mtDNA and mtDNA with a 3.1-kilobase deletion (∆mtDNA) lacking four essential genes5. The heteroplasmic strain, which has 60% ∆mtDNA, displays modest mitochondrial dysfunction and constitutive UPRmt activation. ATFS-1 impairment reduced the ∆mtDNA nearly tenfold, decreasing the total percentage to 7%. We propose that in the context of mtDNA heteroplasmy, UPRmt activation caused by OXPHOS defects propagates or maintains the deleterious mtDNA in an attempt to recover OXPHOS activity by promoting mitochondrial biogenesis and dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bua, E. et al. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 79, 469–480 (2006)
Kraytsberg, Y. et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 38, 518–520 (2006)
Nargund, A. M., Fiorese, C. J., Pellegrino, M. W., Deng, P. & Haynes, C. M. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 58, 123–133 (2015)
Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M. & Haynes, C. M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590 (2012)
Tsang, W. Y. & Lemire, B. D. Stable heteroplasmy but differential inheritance of a large mitochondrial DNA deletion in nematodes. Biochem. Cell Biol. 80, 645–654 (2002)
Greaves, L. C. et al. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 10, e1004620 (2014)
Taylor, S. D. et al. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell 13, 29–38 (2014)
Stewart, J. B. & Chinnery, P. F. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet. 16, 530–542 (2015)
Yoneda, T. et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117, 4055–4066 (2004)
Durieux, J., Wolff, S. & Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011)
Baker, B. M., Nargund, A. M., Sun, T. & Haynes, C. M. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 8, e1002760 (2012)
Haynes, C. M., Petrova, K., Benedetti, C., Yang, Y. & Ron, D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467–480 (2007)
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010)
Valenci, I., Yonai, L., Bar-Yaacov, D., Mishmar, D. & Ben-Zvi, A. Parkin modulates heteroplasmy of truncated mtDNA in Caenorhabditis elegans. Mitochondrion 20, 64–70 (2015)
Suen, D. F., Narendra, D. P., Tanaka, A., Manfredi, G. & Youle, R. J. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl Acad. Sci. USA 107, 11835–11840 (2010)
Scarffe, L. A., Stevens, D. A., Dawson, V. L. & Dawson, T. M. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 37, 315–324 (2014)
Jin, S. M. & Youle, R. J. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 9, 1750–1757 (2013)
Palikaras, K., Lionaki, E. & Tavernarakis, N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 (2015)
Rauthan, M., Ranji, P., Aguilera Pradenas, N., Pitot, C. & Pilon, M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl Acad. Sci. USA 110, 5981–5986 (2013)
Mishra, P., Carelli, V., Manfredi, G. & Chan, D. C. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19, 630–641 (2014)
Chen, H. et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 (2010)
Nakada, K. et al. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 7, 934–940 (2001)
Pellegrino, M. W. et al. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417 (2014)
Schieber, M. & Chandel, N. S. TOR signaling couples oxygen sensing to lifespan in C. elegans. Cell Reports 9, 9–15 (2014)
Lamech, L. T. & Haynes, C. M. The unpredictability of prolonged activation of stress response pathways. J. Cell Biol. 209, 781–787 (2015)
He, L. et al. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res. 30, e68 (2002)
Bargmann, C. I., Hartwieg, E. & Horvitz, H. R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527 (1993)
Andreux, P. A. et al. A method to identify and validate mitochondrial modulators using mammalian cells and the worm C. elegans. Sci. Rep. 4, 5285 (2014)
Acknowledgements
We thank B. Lemire, the National Bioresource Project and the Caenorhabditis Genetics Center for providing C. elegans strains (funded by NIH Office of Research 362 Infrastructure Programs (P40 OD010440), and the Genomics and Bioinformatics Facilities at Memorial Sloan Kettering Cancer Center). This work was supported by National Institutes of Health grants (R01AG040061 and R01AG047182) to C.M.H. and (R01HD078703 and R01NS081490) to S.S., and a Parkinson’s Disease Foundation grant (PDF-FBS-1314) to Y.-F.L. and Deutsche Forschungsgemeinschaft (DFG, SCHU 3023/1-1) to A.M.S.
Author information
Authors and Affiliations
Contributions
Y.-F.L. and C.M.H. planned the experiments. Y.-F.L. generated the worm strains and performed the mtDNA quantification and obtained the images. A.M.S. and Y.-F.L. performed the oxygen consumption analysis and M.W.P. performed the microarray experiments. Electron microscopy was performed by Y.Lu under the supervision of S.S. Y.-F.L. and C.M.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
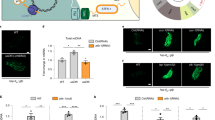
Extended Data Figure 1 RNAi of nuclear encoded OXPHOS components activates the UPRmt, and UPRmt signalling components are required for ∆mtDNA propagation.
a, Images of hsp-6pr::gfp worms raised on nuo-2 (complex I), cyc-1 (cytochrome c) or atp-2 (complex V) RNAi. Scale bar, 0.1 mm. b, Relative ∆mtDNA quantification as determined by qPCR in ∆mtDNA or atfs-1(tm4525);∆mtDNA worms. n = 3; error bars, mean ± s.d.; *P < 0.03 (Student’s t-test). c, Relative ∆mtDNA quantification as determined by qPCR in ∆mtDNA worms raised on control, dve-1, or ubl-5 RNAi. n = 3; error bars, mean ± s.d.; *P < 0.03 (Student’s t-test). d, Images of dve-1pr::dve-1::gfp in wild-type or ∆mtDNA worms. Scale bar, 0.1 mm. e, Ratios of ∆mtDNA and nuclear genomic DNA as determined by qPCR in individual ∆mtDNA and atfs-1(tm4919);∆mtDNA worms. Black bars represent the mean. n = 16; mean P < 0.0001 (Student’s t-test).
Extended Data Figure 2 The reduction in ∆mtDNA caused by atfs-1(RNAi) is largely independent of autophagy, and ATFS-1 activation in the presence of a deleterious mtDNA is harmful.
a, Relative ∆mtDNA quantification as determined by qPCR in ∆mtDNA and atg-18(gk378);∆mtDNA worms raised on control or atfs-1 RNAi. n = 3; error bars, mean ± s.d.; *P < 0.03 (Student’s t-test). b, Images of hsp-6pr::gfp worms raised on control or spg-7 RNAi. Scale bar, 0.1 mm. c, Images of TMRE-stained wild-type, ∆mtDNA, atfs-1(et18) and atfs-1(et18);∆mtDNA worms. Scale bar, 0.1 mm. d, Images of synchronized atfs-1(et18) and atfs-1(et18);∆mtDNA worms raised on 2.5 μM rotenone 4 days after hatching. Scale bar, 1 mm.
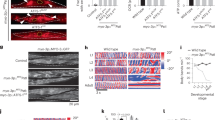
Extended Data Figure 3 polg-1, hmg-5 and drp-1 mRNAs are induced in atfs-1(et18) worms, and mitochondrial fusion is required for the development of atfs-1(et18) worms harbouring ∆mtDNA.
a, polg-1, hmg-5 and drp-1 transcripts as determined by qRT–PCR in wild-type and atfs-1(et18) worms. n = 3; error bars, mean ± s.d.; *P < 0.03 (Student’s t-test). RFU, random fluorescent units. b, Images of synchronized ∆mtDNA, atfs-1(et18), and atfs-1(et18);∆mtDNA worms raised on control or fzo-1 RNAi. Scale bar, 1 mm. c, ∆mtDNA quantification as determined by qPCR in ∆mtDNA worms raised on control or hmg-5 RNAi. n = 3; error bars, mean ± s.d.; *P < 0.03 (Student’s t-test).
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-2 comprising: (1) the relative fold induction and P values for each mRNA and (2) list of Primers that specifically amplify wild-type or ΔmtDNA . (PDF 242 kb)
Rights and permissions
About this article
Cite this article
Lin, YF., Schulz, A., Pellegrino, M. et al. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533, 416–419 (2016). https://doi.org/10.1038/nature17989
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17989
This article is cited by
-
Mitochondrial stress: a key role of neuroinflammation in stroke
Journal of Neuroinflammation (2024)
-
Coupling of autophagy and the mitochondrial intrinsic apoptosis pathway modulates proteostasis and ageing in Caenorhabditis elegans
Cell Death & Disease (2023)
-
MRPS6 modulates glucose-stimulated insulin secretion in mouse islet cells through mitochondrial unfolded protein response
Scientific Reports (2023)
-
ATFS-1 counteracts mitochondrial DNA damage by promoting repair over transcription
Nature Cell Biology (2023)
-
Insight into the mitochondrial unfolded protein response and cancer: opportunities and challenges
Cell & Bioscience (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.