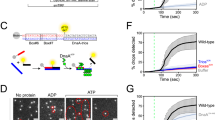
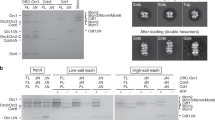
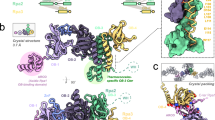
Abstract
DNA replication is tightly controlled to ensure accurate inheritance of genetic information. In all organisms, initiator proteins possessing AAA+ (ATPases associated with various cellular activities) domains bind replication origins to license new rounds of DNA synthesis1. In bacteria the master initiator protein, DnaA, is highly conserved and has two crucial DNA binding activities2. DnaA monomers recognize the replication origin (oriC) by binding double-stranded DNA sequences (DnaA-boxes); subsequently, DnaA filaments assemble and promote duplex unwinding by engaging and stretching a single DNA strand3,4,5. While the specificity for duplex DnaA-boxes by DnaA has been appreciated for over 30 years, the sequence specificity for single-strand DNA binding has remained unknown. Here we identify a new indispensable bacterial replication origin element composed of a repeating trinucleotide motif that we term the DnaA-trio. We show that the function of the DnaA-trio is to stabilize DnaA filaments on a single DNA strand, thus providing essential precision to this binding mechanism. Bioinformatic analysis detects DnaA-trios in replication origins throughout the bacterial kingdom, indicating that this element is part of the core oriC structure. The discovery and characterization of the novel DnaA-trio extends our fundamental understanding of bacterial DNA replication initiation, and because of the conserved structure of AAA+ initiator proteins these findings raise the possibility of specific recognition motifs within replication origins of higher organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Duderstadt, K. E. & Berger, J. M. A structural framework for replication origin opening by AAA + initiation factors. Curr. Opin. Struct. Biol. 23, 144–153 (2013)
Messer, W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26, 355–374 (2002)
Fuller, R. S., Funnell, B. E. & Kornberg, A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38, 889–900 (1984)
Duderstadt, K. E., Chuang, K. & Berger, J. M. DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213 (2011)
Fujikawa, N. et al. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086 (2003)
Mackiewicz, P., Zakrzewska-Czerwinska, J., Zawilak, A., Dudek, M. R. & Cebrat, S. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res. 32, 3781–3791 (2004)
Wolański, M., Donczew, R., Zawilak-Pawlik, A. & Zakrzewska-Czerwińska, J. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 5, 735 (2015)
Hassan, A. K. et al. Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J. Bacteriol. 179, 2494–2502 (1997)
Krause, M., Rückert, B., Lurz, R. & Messer, W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274, 365–380 (1997)
Leonard, A. C. & Grimwade, J. E. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65, 19–35 (2011)
Speck, C. & Messer, W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20, 1469–1476 (2001)
Scholefield, G., Errington, J. & Murray, H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31, 1542–1555 (2012)
Cheng, H. M., Gröger, P., Hartmann, A. & Schlierf, M. Bacterial initiators form dynamic filaments on single-stranded DNA monomer by monomer. Nucleic Acids Res. 43, 396–405 (2015)
Ozaki, S. et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362 (2008)
Gaudier, M., Schuwirth, B. S., Westcott, S. L. & Wigley, D. B. Structural basis of DNA replication origin recognition by an ORC protein. Science 317, 1213–1216 (2007)
Dueber, E. L., Corn, J. E., Bell, S. D. & Berger, J. M. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317, 1210–1213 (2007)
Erzberger, J. P., Mott, M. L. & Berger, J. M. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nature Struct. Mol. Biol. 13, 676–683 (2006)
Duderstadt, K. E. et al. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 285, 28229–28239 (2010)
Krause, M. & Messer, W. DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene 228, 123–132 (1999)
Donczew, R., Weigel, C., Lurz, R., Zakrzewska-Czerwinska, J. & Zawilak-Pawlik, A. Helicobacter pylori oriC – the first bipartite origin of chromosome replication in Gram-negative bacteria. Nucleic Acids Res. 40, 9647–9660 (2012)
Ozaki, S., Fujimitsu, K., Kurumizaka, H. & Katayama, T. The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima forms an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells 11, 425–438 (2006)
Gao, F., Luo, H. & Zhang, C. T. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41, D90–D93 (2013)
Kumar, S., Farhana, A. & Hasnain, S. E. In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PLoS ONE 4, e4139 (2009)
Pei, H. et al. Mechanism for the TtDnaA-Tt-oriC cooperative interaction at high temperature and duplex opening at an unusual AT-rich region in Thermoanaerobacter tengcongensis. Nucleic Acids Res. 35, 3087–3099 (2007)
Kaur, G. et al. Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 91, 1148–1163 (2014)
Noguchi, Y., Sakiyama, Y., Kawakami, H. & Katayama, T. The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 290, 20295–20312 (2015)
Bleichert, F., Botchan, M. R. & Berger, J. M. Crystal structure of the eukaryotic origin recognition complex. Nature 519, 321–326 (2015)
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004)
Taylor, R. G., Walker, D. C. & McInnes, R. R. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678 (1993)
Morimoto, T. et al. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148, 3539–3552 (2002)
Vagner, V., Dervyn, E. & Ehrlich, S. D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144, 3097–3104 (1998)
Berkmen, M. B. & Grossman, A. D. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol. Microbiol. 63, 150–165 (2007)
Hansen, E. B. & Yarmolinsky, M. B. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc. Natl Acad. Sci. USA 83, 4423–4427 (1986)
Moriya, S., Atlung, T., Hansen, F. G., Yoshikawa, H. & Ogasawara, N. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol. 6, 309–315 (1992)
Oka, A., Sugimoto, K., Takanami, M. & Hirota, Y. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol. Gen. Genet. 178, 9–20 (1980)
Calcutt, M. J. & Schmidt, F. J. Conserved gene arrangement in the origin region of the Streptomyces coelicolor chromosome. J. Bacteriol. 174, 3220–3226 (1992)
Watanabe, S. et al. Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol. Microbiol. 83, 856–865 (2012)
Acknowledgements
We thank J. Errington and W. Vollmer for reviewing the manuscript. We thank G. Scholefield for preliminary data, A. Koh for research assistance and I. Selmes for technical assistance. Research support was provided to H.M. by a Royal Society University Research Fellowship and a Biotechnology and Biological Sciences Research Council Research Grant (BB/K017527/1), and to O.H. by an Iraqi Ministry of Higher Education and Scientific Research Studentship.
Author information
Authors and Affiliations
Contributions
H.M. and T.T.R. conceived and designed experiments; H.M., T.T.R. and O.H. constructed plasmids and strains; H.M. and O.H. performed growth and marker frequency analysis experiments; H.M. performed microscopy experiments; T.T.R. purified proteins, performed the open complex assay, and performed the DnaA filament formation assays; H.M. and T.T.R. interpreted results and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Structure of DnaA proteins.
a, Primary domain structure of DnaA. Key functions are listed below the relevant domain. b, Structure of Thermatoga maritima DnaA domain III, highlighting the single-strand binding residue Val176 (Ile190 B. subtilis) within the ISM (PDB accession number 2Z4S). c, Structure of E. coli DnaA domain IV bound to a DnaA-box (PDB accession number 1J1V). d, Structure of A. aeolicus DnaA domain III (blue shades) and domain IV (cyan shades) bound to a single DNA strand (orange), highlighting the single-strand binding residue Val156 (Ile190 B. subtilis) (PDB accession number 3R8F). e, Scheme used to construct mutants within the B. subtilis DNA replication origin. The green arrow highlights the location of a DnaA-box mutation.
Extended Data Figure 2 Characterization of the inducible repN/oriN replication initiation system.
Repression of repN expression inhibits DNA replication in a ΔoriC mutant. A large deletion was introduced into the B. subtilis replication origin using a strain harbouring the inducible oriN/repN construct. Strain growth was found to be dependent upon addition of the inducer IPTG. a, Strains streaked to resolve single colonies. b, A GFP-DnaN reporter was used to detect DNA replication after removal of IPTG from inducible oriN/repN strains. Scale bar, 5 μm. c, Genetic map indicating the location of oriN at the aprE locus in strain HM1108. d, Analysis of DNA replication initiation at oriC and oriN. Marker frequency analysis was used to measure the rate of DNA replication initiation in the presence and absence of IPTG (0.1 mM). Genomic DNA was harvested from cells during the exponential growth phase and the relative amount of DNA from either the endogenous replication origin (oriC) or the aprE locus (oriN) compared with the terminus (ter) was determined using qPCR (mean and s.d. of three technical replicates). Cell doubling times (in minutes) are shown above each data set.
Extended Data Figure 3 Wild-type DnaA assembles into filaments on 5′-tailed substrates.
DnaA filament formation using amine-specific crosslinking (BS3) on DNA scaffolds (represented by symbols above each lane). Protein complexes were resolved by SDS–PAGE and DnaA was detected by western blot analysis.
Extended Data Figure 4 DNA sequence of unwinding regions after mononucleotide and trinucleotide deletions.
Resulting sequences grouped in boxes are identical for more than one deletion.
Extended Data Figure 5 Crosslinking with BS3 captures a distinct DnaA oligomer.
DnaA was incubated with various DNA scaffolds and different crosslinking agents were added to capture distinct DnaA oligomers. a, Crosslinking with BMOE detects DnaA oligomers forming on both duplex and tailed substrates. b, Crosslinking with BS3 only detects DnaA oligomers forming on tailed substrates, revealing an interaction between DnaA and the first DnaA-trio motif located downstream of the GC-cluster.
Extended Data Figure 6 The nucleotide at the third position of the DnaA-trio is required to stabilize DnaA.
DNA scaffolds containing the first two nucleotides of a DnaA-trio either with or without a 5′-phosphate are unable to stabilize binding of an additional DnaA protomer, indicating that the nucleotide at the third position is required. Combined with the data shown in Fig. 4b where the position is abasic, the results suggest that the sugar at the third position plays a critical role in DnaA binding.
Extended Data Figure 7 Relationship between the DnaA-box and the DnaA-trios.
a, Sequence of the origin region used for constructing DNA scaffolds. Symbols below represent duplex DnaA-boxes (triangles), the GC-rich region (green rectangles), the two strands of the unwinding region (red or pink rectangles) and the AT-rich region (blue rectangle). b, Loading of the DnaA filament onto a single-stranded 5′-tail requires a DnaA-box and DnaA domains III–IV, but the DnaA-box position and orientation are flexible.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Tables 1-2 and additional references. (PDF 1000 kb)
Rights and permissions
About this article
Cite this article
Richardson, T., Harran, O. & Murray, H. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature 534, 412–416 (2016). https://doi.org/10.1038/nature17962
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17962
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.