Abstract
Asymmetric cell division, the partitioning of cellular components in response to polarizing cues during mitosis, has roles in differentiation and development1. It is important for the self-renewal of fertilized zygotes in Caenorhabditis elegans and neuroblasts in Drosophila, and in the development of mammalian nervous and digestive systems1. T lymphocytes, upon activation by antigen-presenting cells (APCs), can undergo asymmetric cell division, wherein the daughter cell proximal to the APC is more likely to differentiate into an effector-like T cell and the distal daughter is more likely to differentiate into a memory-like T cell2. Upon activation and before cell division, expression of the transcription factor c-Myc drives metabolic reprogramming, necessary for the subsequent proliferative burst3. Here we find that during the first division of an activated T cell in mice, c-Myc can sort asymmetrically. Asymmetric distribution of amino acid transporters, amino acid content, and activity of mammalian target of rapamycin complex 1 (mTORC1) is correlated with c-Myc expression, and both amino acids and mTORC1 activity sustain the differences in c-Myc expression in one daughter cell compared to the other. Asymmetric c-Myc levels in daughter T cells affect proliferation, metabolism, and differentiation, and these effects are altered by experimental manipulation of mTORC1 activity or c-Myc expression. Therefore, metabolic signalling pathways cooperate with transcription programs to maintain differential cell fates following asymmetric T-cell division.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
20 April 2016
The text of the first paragraph has been corrected.
References
Pham, K., Sacirbegovic, F. & Russell, S. M. Polarised cells, polarised views: asymmetric cell division in hematopoietic cells. Front. Immunol. 5, (2014)
Chang, J. T. et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687–1691 (2007)
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011)
Huang, C. Y., Bredemeyer, A. L., Walker, L. M., Bassing, C. H. & Sleckman, B. P. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 38, 342–349 (2008)
Colombetti, S., Basso, V., Mueller, D. L. & Mondino, A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J. Immunol. 176, 2730–2738 (2006)
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008)
Grohmann, U. & Bronte, V. Control of immune response by amino acid metabolism. Immunol. Rev. 236, 243–264 (2010)
Choo, A. Y., Yoon, S. O., Kim, S. G., Roux, P. P. & Blenis, J. Rapamycin differentially inhibits S6Ks and 4E–BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl Acad. Sci. USA 105, 17414–17419 (2008)
Hsu, P. P. et al. The mTOR-Regulated Phosphoproteome Reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011)
Ouyang, W., Beckett, O., Flavell, R. A. & Li, M. O. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009)
Peck, B., Ferber, E. C. & Schulze, A. Antagonism between FOXO and Myc regulates cellular powerhouse. Front. Oncol. 3, (2013)
Yang, K., Neale, G., Green, D. R., He, W. & Chi, H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nature Immunol. 12, 888–897 (2011)
Vervoorts, J., Luscher-Firzlaff, J. & Luscher, B. The ins and outs of MYC regulation by posttranslational mechanisms. J. Biol. Chem. 281, 34725–34729 (2006)
Sears, R. C. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3, 1133–1137 (2004)
Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA 105, 18782–18787 (2008)
Wang, R. & Green, D. R. Metabolic checkpoints in activated T cells. Nature Immunol. 13, 907–915 (2012)
Marelli-Berg, F. M., Fu, H. & Mauro, C. Molecular mechanisms of metabolic reprogramming in proliferating cells: implications for T-cell-mediated immunity. Immunology 136, 363–369 (2012)
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012)
van der Windt, G. J. & Pearce, E. L. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol. Rev. 249, 27–42 (2012)
Katajisto, P. et al. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343 (2015)
Arsenio, J. et al. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nature Immunol. 15, 365–372 (2014)
Stemberger, C. et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity 27, 985–997 (2007)
Schepers, K. et al. Dissecting T cell lineage relationships by cellular barcoding. J. Exp. Med. 205, 2309–2318 (2008)
Bannard, O., Kraman, M. & Fearon, D. T. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science 323, 505–509 (2009)
Harrington, L. E., Janowski, K. M., Oliver, J. R., Zajac, A. J. & Weaver, C. T. Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452, 356–360 10.1038/nature06672 (2008)
Rohr, J. C., Gerlach, C., Kok, L. & Schumacher, T. N. Single cell behavior in T cell differentiation. Trends Immunol. 35, 170–177 (2014)
Dang, C. V., Le, A. & Gao, P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15, 6479–6483 (2009)
Gao, P. et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009)
Liu, J. & Levens, D. Making myc. Curr. Top. Microbiol. Immunol. 302, 1–32 (2006)
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009)
Acknowledgements
We thank R. Cross, G. Lennon, and P. Ingle for cell sorting, P. Thomas for assistance with influenza infections, H. Chi for help with Listeria infections, and I. Pawlikowska for help with statistical analyses. This work was supported by ALSAC and grants from the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
K.C.V. conceived the project, designed and performed most experiments, interpreted results, and co-wrote the manuscript. C.S.G. guided imaging, performed live imaging experiments, and performed experiments with TSC1 flox/flox animals. S.M. provided technical assistance with Seahorse XF24 assays and helped with several in vivo studies. S.L. performed some imaging experiments with amino acid transports, performed experiments with high IL-2, and helped with several in vivo studies. M.M.K. performed and analysed all gene expression analyses. R.W. provided intellectual contributions. D.R.G. conceived the project, supervised experimental designs, interpreted results, and co-wrote the manuscript.
Corresponding author
Extended data figures and tables
Extended Data Figure 1 Characterization of c-Myc asymmetry.
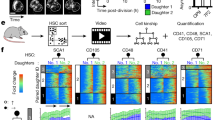
a, Gating strategy for CD8high and CD8low cells. CellTrace-Violet-labelled naive T cells were activated with anti-CD3, anti-CD28, and ICAM for 36 h. CD8 high or low cells were identified as activated, undivided (U), first division (1), or second division (2) on the basis of dilution of CellTrace Violet. Frequencies of cells in each population are presented in table below. b, Time course of CellTrace Violet dilution in naive, T cells activated with anti-CD3, anti-CD28, and ICAM for the indicated time points. c, Schematic representation of staining patterns for interpretation of data represented in Fig. 1c–h. Black lines indicate axes on graphs. d, Frequencies of asymmetric cell division were determined by analysing 92 conjoined daughter cells from cultures activated with anti-CD3, anti-CD28, and ICAM and 123 conjoined daughter cells from cultures activated with SIINFEKL-pulsed BMDCs. T cells were analysed for c-Myc–GFP intensity in both daughter cells. Using 1.5-times brightness in one daughter versus the other as a cut off, each pair was assigned either asymmetric or not, and the frequency of asymmetric pairs is plotted.
Extended Data Figure 2 Representative confocal images and single stains.
For data in Fig. 1. a, Fig. 1b, c; b, Fig. 1d, e; c, Fig. 1f; d, Fig. 1g; e, Fig. 1h.
Extended Data Figure 3 c-Myc expression becomes asymmetric in late telophase.
Overlay, β-tubulin (blue), Hoechst 33258 (grey), c-Myc–GFP (green), and phalloidin (red) for each mitotic phase indicated on the left as identified by chromatin and tubulin staining patterns.
Extended Data Figure 4 Asymmetric assortment of mRNA for c-Myc target genes in activated T cells.
CellTrace-Violet-labelled T cells were stimulated for 36 h with anti-CD3, anti-CD28, and ICAM. a–c, First division (a, c) or undivided (a–c) c-Mychigh (H) and c-Myclow (L) T cells were sorted on the basis of c-Myc–GFP expression (b is undivided cells only), and RNA was extracted from each population. Fold change in gene expression for the indicated primers is quantified using the 2−ΔΔCt method relative to β-2-microglobulin across three (a, c) or four (b) independent experiments (mean ± s.d.).
Extended Data Figure 5 Cytokine signalling does not influence c-Myc asymmetry and asymmetric assortment of CD98.
a, Mean fluorescent intensity (MFI) of c-Myc–GFP (left panel) or CD98 (right panel) in CD8high (shaded bars) and CD8low (open bars) in the first division after 35 h of activation by anti-CD3, anti-CD8, and ICAM and the indicated treatment for 1 h. All differences were determined significant (P < 0.05) by unpaired Student’s t-test. b, Flow cytometric analysis of c-Myc GFP and CD98 in CD8high (green histograms), CD8low (grey histograms), or IL-7 (5 ng ml−1) rested unactivated (gold histograms) T cells in control conditions or activated in the presence of 10 ng ml−1 IL-2 (0–36 h after activation). Representative flow plots are on the left, and quantification of the mean fluorescence intensities of c-Myc and CD98-APC are on the right. Experiment representative of three independent experiments. c, Representative confocal image and single stain images for Fig. 2d, e. OT-I T cells stimulated on SIINFEKL-pulsed BMDCs d, Quantification of CD98 and c-Myc asymmetry in T cells stimulated with anti-CD3, anti-CD28, and ICAM for 36 h. 100% concordance of markers (P = 0.0039 Two-Tailed Binomial Test); R2 = 0.2304, P = 0.2304 linear regression. e, Polarization of SLC3A2 in two representative confocal images in antibody-stimulated OT-I Tg T cells (see methods). f, Asymmetric index (difference in RFP Intensity in proximal (P) and distal (D) sides of cell/sum of RFP intensities in proximal and distal sides of cell) for SLC3A2 staining in activated, undivided CD8 T cells (each point represents an activated T cell). Error bars, mean ± s.d.
Extended Data Figure 6 Amino acid transporter SLC1A5, but not SLC1A3, asymmetrically assorts in activated T cells.
a, b, Representative confocal image, quantification, and single-stain images for SLC1A5; 88.9% both bright in proximal daughter (x2 = 19.89, DF = 3, P = 0.0002, chi-square goodness of fit test); R2 = 0.2961, P = 0.1299, linear regression (a); and SLC1A3 100% c-Myc bright in proximal daughter, 62.5% SLC1A3 bright in distal daughter (x2 = 9, DF = 3, P = 0.0293 chi-square goodness of fit test); R2 = 0.07944, P = 0.4989 linear regression (b) for OT-I CD8 T cells co-cultured with SIINFEKL-pulsed BMDCs for 36 h.
Extended Data Figure 7 Regulation of p-mTOR and mTORC1 signalling by c-Myc.
a, b, Quantification of p-mTOR staining and c-Myc–GFP for OT-I CD8 T cells co-cultured with SIINFEKL-pulsed BMDCs 80.9% both bright in proximal daughter (x2 = 25.67, DF = 3, P < 0.0001 chi-square goodness of fit test); R2 = 0.2307, P = 0.0275, linear regression (a); or T cells stimulated for 36 h with anti-CD3, anti-CD28, and ICAM 82.4% concordance of markers (P = 0.0013, two-tailed binomial test); R2 = 0.1204, P = 0.1725, linear regression (b). Asymmetry as assessed by fluorescence intensity is expressed as (proximal − distal)/total (a), or values from (c-Mychigh – c-Myclow)/total (b). c, Western blot analysis of CD8 T cells activated with anti-CD3, anti-CD28, and ICAM for 6 h without treatment (ctrl) or with 1 μM or 5 μM of the bromodomain inhibitor JQ1. Data are representative of three independent experiments.
Extended Data Figure 8 Asymmetric assortment of mTORC1 activity with c-Myc–GFP.
a, b, Representative confocal images for quantifications of c-Myc–GFP and pS6 staining corresponding to Fig. 3a, b (a) or c-Myc–GFP and p70S6K corresponding to Fig. 3c (b) in OT-I T cells co-cultured with SIINFEKL-pulsed BMDCs. c, d, Quantifications for c-Myc–GFP and pS6 100% both bright in proximal daughter (x2 = 19.89, DF = 3, P = 0.0002, chi-square goodness of fit test); R2 = 0.9457, P = 0.0055, linear regression (c); or p70S6K 100% both bright in proximal daughter (x2 = 14.14, DF = 3, P = 0.0027, chi-square goodness of fit test); R2 = 0.4875, P = 0.0026, linear regression (d) staining in T cells stimulated on anti-CD3, anti-CD28, and ICAM for 36 h. e, Representative confocal images for quantifications of c-Myc–GFP and pFOXO1 staining corresponding to Fig. 3d, e. T cells stimulated with anti-CD3, anti-CD28, and ICAM for 36 h.
Extended Data Figure 9 First-division CD8high c-Myc–GFPhigh CD8 T cells are more glycolytic and exhibit more glutaminolysis and pentose phosphate pathway activity than first division CD8low c-Myc–GFPlow CD8 T cells but have decreased FAO.
Undivided or first-division antibody-activated T cells were sorted on c-Myc expression. 1.6 million T cells from each group were analysed via UHPLC/MS/MS for unbiased metabolomic profiling performed by Metabolon, Inc. a–d, Metabolic pathway schematic (a) was generated by Metabolon Inc., as were graphs (b) for select metabolites in pathway, representing data for DNA-normalized data. Metabolic pathway schematics and graphs were also generated by Metabolon Inc. for select metabolites in the pathway, representing data for DNA-normalized data for nucelotide biosynthesis (c) and fatty acid oxidation (d).
Extended Data Figure 10 c-Mychigh CD8 T cells are more glycolytic than CD8low CD8 T cells without asymmetric distribution of mitochondria.
a, The extracellular acidification rate (ECAR) for sorted c-Mychigh (circles) and c-Myclow (squares) T cells from the first division after 36 h of activation with anti-CD3, anti-CD28, and ICAM as measured by a Seahorse Bioflux analyser during exposure to the indicated compounds are represented. Values are paired across three independent experiments. b, The oxygen consumption rate (top panels) and ECAR (bottom panels) for basal respiration in complete RPMI with glucose of sorted c-Mychigh (shaded bars) and c-Myclow (open bars) CD8 T cells from the first-division after 36 h of activation by anti-CD3, anti-CD28, and ICAM, as measured by a Seahorse Bioflux analyser across three independent experiments. c, Quantification of mitochondrial DNA in sorted first-division c-Myc low and high OT-I CD8 T cells activated on anti-CD3, anti-CD28 and ICAM for 36 h. Mean + s.d. are plotted for three technical replicates of n = 2 mice per group. d, Overlay and individual channel images of β-tubulin (blue), DAPI (grey), c-Myc–GFP (green), and TOM20 (red) of an OT-I CD8 T cell activated with anti-CD3, anti-CD28 and ICAM for 36 h.
Supplementary information
Supplementary Information
This file contains additional Supplementary Methods and References. (PDF 213 kb)
Asymmetric assortment of c-Myc in a dividing CD8+ T cell.
T cells from c-Myc-GFP knockin mice were stimulated by culture on plate bound anti-CD3, anti-CD28, and ICAM (see methods), then fixed. Shown is a 3D reconstruction turned about the x-axis of volumetric representations of compiled images taken at 0.4 microns. Beta tubulin is pseudo-coloured blue, c-Myc-GFP green, and CD8 red. (MP4 119 kb)
Asymmetric assortment of c-Myc in a dividing CD8+ T cell.
T cells from c-Myc-GFP knockin mice were stimulated by culture on plate bound anti-CD3, anti-CD28, and ICAM (see methods), then fixed. Shown is a 3D reconstruction turned about the x-axis of volumetric representations of compiled images taken at 0.4 microns. Beta tubulin is pseudo-coloured blue, c-Myc-GFP green, and CD8 red. (MP4 90 kb)
Asymmetric assortment of c-Myc in a dividing CD8+ T cell.
T cells from c-Myc-GFP knockin mice were stimulated by culture on plate bound anti-CD3, anti-CD28, and ICAM (see methods), then fixed. Shown is a 3D reconstruction turned about the x-axis of volumetric representations of compiled images taken at 0.4 microns. Beta tubulin is pseudo-coloured blue, c-Myc-GFP green, and CD8 red. (MP4 124 kb)
Asymmetric assortment of c-Myc in a dividing CD8+ T cell.
OT-I, c-Myc-GFP knockin T cells were stimulated by culture with SIINFEKL-pulsed BMDC (see methods), then fixed. Shown is a 3D reconstruction turned about the x-axis of volumetric representations of compiled images taken at 0.4 microns. Beta tubulin is pseudo-coloured blue, c-Myc-GFP green, and CD8 red. (MP4 199 kb)
Asymmetric assortment of c-Myc in a dividing CD8+ T cell.
OT-I, c-Myc-GFP knockin T cells were stimulated by culture with SIINFEKL-pulsed BMDC (see methods), then fixed. Shown is a 3D reconstruction turned about the x-axis of volumetric representations of compiled images taken at 0.4 microns. Beta tubulin is pseudo-coloured blue, c-Myc-GFP green, and CD8 red. (MP4 163 kb)
Asymmetric assortment of c-Myc in a dividing CD8+ T cell
OT-I, c-Myc-GFP knockin T cells were stimulated by culture with SIINFEKL-pulsed BMDC (see methods), then fixed. Shown is a 3D reconstruction turned about the x-axis of volumetric representations of compiled images taken at 0.4 microns. Beta tubulin is pseudo-coloured blue, c-Myc-GFP green, and CD8 red. (MP4 229 kb)
Time-lapse of asymmetric assortment of c-Myc in a dividing CD8+ T cell.
Compilation of images taken every 2 minutes of a dividing OT-I, c-Myc-GFP knockin T cell after co-culture with SIINFEKL-pulsed BMDCs. Cell Trace Violet, blue; c-Myc-GFP, green. Tiles in Figure 2A were taken from every other still in this compilation. (MP4 402 kb)
Rights and permissions
About this article
Cite this article
Verbist, K., Guy, C., Milasta, S. et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393 (2016). https://doi.org/10.1038/nature17442
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17442
This article is cited by
-
Amino acid metabolism in tumor biology and therapy
Cell Death & Disease (2024)
-
Amino acid metabolism in immune cells: essential regulators of the effector functions, and promising opportunities to enhance cancer immunotherapy
Journal of Hematology & Oncology (2023)
-
Regulation of CD8+ T memory and exhaustion by the mTOR signals
Cellular & Molecular Immunology (2023)
-
c-Myc uses Cul4b to preserve genome integrity and promote antiviral CD8+ T cell immunity
Nature Communications (2023)
-
Radiotherapy remodels the tumor microenvironment for enhancing immunotherapeutic sensitivity
Cell Death & Disease (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.