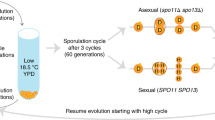
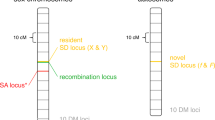
Abstract
Sex and recombination are pervasive throughout nature despite their substantial costs1. Understanding the evolutionary forces that maintain these phenomena is a central challenge in biology2,3. One longstanding hypothesis argues that sex is beneficial because recombination speeds adaptation4. Theory has proposed several distinct population genetic mechanisms that could underlie this advantage. For example, sex can promote the fixation of beneficial mutations either by alleviating interference competition (the Fisher–Muller effect)5,6 or by separating them from deleterious load (the ruby in the rubbish effect)7,8. Previous experiments confirm that sex can increase the rate of adaptation9,10,11,12,13,14,15,16,17, but these studies did not observe the evolutionary dynamics that drive this effect at the genomic level. Here we present the first, to our knowledge, comparison between the sequence-level dynamics of adaptation in experimental sexual and asexual Saccharomyces cerevisiae populations, which allows us to identify the specific mechanisms by which sex speeds adaptation. We find that sex alters the molecular signatures of evolution by changing the spectrum of mutations that fix, and confirm theoretical predictions that it does so by alleviating clonal interference. We also show that substantially deleterious mutations hitchhike to fixation in adapting asexual populations. In contrast, recombination prevents such mutations from fixing. Our results demonstrate that sex both speeds adaptation and alters its molecular signature by allowing natural selection to more efficiently sort beneficial from deleterious mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Bell, G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality (Univ. California Press, 1982)
Otto, S. P. & Lenormand, T. Resolving the paradox of sex and recombination. Nature Rev. Genet. 3, 252–261 (2002)
Kondrashov, A. S. Classification of hypotheses on the advantage of amphimixis. J. Hered. 84, 372–387 (1993)
Weismann, A. in Essays upon Heredity and Kindred Biological Problems (eds Poulton, E. B., Schonland, S. & Shipley, A. E. ) 251–332 (Clarendon, 1889)
Fisher, R. A. The Genetical Theory of Natural Selection Ch. 6 (Oxford Univ. Press, 1930)
Muller, H. Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932)
Peck, J. R. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137, 597–606 (1994)
Johnson, T. & Barton, N. H. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162, 395–411 (2002)
Gray, J. C. & Goddard, M. R. Sex enhances adaptation by unlinking beneficial from detrimental mutations in experimental yeast populations. BMC Evol. Biol. 12, 43 (2012)
Becks, L. & Agrawal, A. F. The evolution of sex is favoured during adaptation to new environments. PLoS Biol. 10, e1001317 (2012)
Zeyl, C. & Bell, G. The advantage of sex in evolving yeast populations. Nature 388, 465–468 (1997)
Goddard, M. R., Godfray, H. C. J. & Burt, A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434, 636–640 (2005)
Colegrave, N. Sex releases the speed limit on evolution. Nature 420, 664–666 (2002)
Poon, A. & Chao, L. Drift increases the advantage of sex in RNA bacteriophage Φ6. Genetics 166, 19–24 (2004)
Becks, L. & Agrawal, A. F. Higher rates of sex evolve in spatially heterogeneous environments. Nature 468, 89–92 (2010)
Rice, W. R. & Chippindale, A. K. Sexual recombination and the power of natural selection. Science 294, 555–559 (2001)
Cooper, T. F. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biol. 5, e225 (2007)
Weissman, D. B. & Barton, N. H. Limits to the rate of adaptive substitution in sexual populations. PLoS Genet. 8, e1002740 (2012)
Crow, J. F. & Kimura, M. Evolution in sexual and asexual populations. Am. Nat. 99, 439–450 (1965)
Maynard Smith, J. What use is sex? J. Theor. Biol. 30, 319–335 (1971)
Lang, G. I. et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500, 571–574 (2013)
Kao, K. C. & Sherlock, G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature Genet. 40, 1499–1504 (2008)
Miralles, R., Gerrish, P. J., Moya, A. & Elena, S. F. Clonal interference and the evolution of RNA viruses. Science 285, 1745–1747 (1999)
Sella, G., Petrov, D. A., Przeworski, M. & Andolfatto, P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5, e1000495 (2009)
Good, B. H. & Desai, M. M. Deleterious passengers in adapting populations. Genetics 198, 1183–1208 (2014)
Schiffels, S., Szöllősi, G. J., Mustonen, V. & Lässig, M. Emergent neutrality in adaptive asexual evolution. Genetics 189, 1361–1375 (2011)
Kondrashov, A. S. Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440 (1988)
Hartfield, M. & Otto, S. P. Recombination and hitchhiking of deleterious alleles. Evolution 65, 2421–2434 (2011)
Birky, C. W. & Walsh, J. B. Effects of linkage on rates of molecular evolution. Proc. Natl Acad. Sci. USA 85, 6414–6418 (1988)
Frenkel, E. M. et al. Crowded growth leads to the spontaneous evolution of semi-stable coexistence in laboratory yeast populations. Proc. Natl Acad. Sci. USA 112, 11306–11311 (2015)
Tong, A. H. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 (2001)
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002)
Lang, G. I., Botstein, D. & Desai, M. M. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188, 647–661 (2011)
Hall, B. M., Ma, C.-X., Liang, P. & Singh, K. K. Fluctuation AnaLysis CalculatOR: a web tool for the determination of mutation rate using Luria–Delbrück fluctuation analysis. Bioinformatics 25, 1564–1565 (2009)
Frenkel, E. M., Good, B. H. & Desai, M. M. The fates of mutant lineages and the distribution of fitness effects of beneficial mutations in laboratory budding yeast populations. Genetics 196, 1217–1226 (2014)
Kryazhimskiy, S., Rice, D. P., Jerison, E. R. & Desai, M. M. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344, 1519–1522 (2014)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012)
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498 (2011)
Anscombe, F. J. The transformation of poisson, binomial and negative-binomial data. Biometrika 35, 246–254 (1948)
Donoho, D. L. & Johnstone, I. M. Adapting to unknown smoothness via wavelet shrinkage. J. Am. Stat. Assoc. 90, 1200–1224 (1995)
Shim, H. & Stephens, M. Wavelet-based genetic association analysis of functional phenotypes arising from high-throughput sequencing assays. Ann. Appl. Stat. 9, 665–686 (2015)
Dunham, M. J. et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 99, 16144–16149 (2002)
Chang, S.-L., Lai, H.-Y., Tung, S.-Y. & Leu, J.-Y. Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS Genet. 9, e1003232 (2013)
Acknowledgements
We thank J.-Y. Leu and S. Akle-Serrano for help with strain construction and experimental evolution; S. Kryazhimskiy, E. Jerison, and J. Piper for help with sequencing library preparation; G. Lang, A. Murray, B. Good, D. van Dyken, K. Kosheleva, I. Cvijovic´, and other members of the Desai laboratory for discussions and comments on the manuscript; and P. Rogers and C. Daly for technical support. D.P.R. acknowledges support from an NSF graduate research fellowship. M.M.D. acknowledges support from the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, the Harvard Milton Fund, the Simons Foundation (grant 376196), grant PHY 1313638 from the National Science Foundation, and grant GM104239 from the National Institutes of Health. Computational work was performed on the Odyssey cluster supported by the Research Computing Group at Harvard University.
Author information
Authors and Affiliations
Contributions
M.J.M., D.P.R., and M.M.D. designed the project; M.J.M. conducted the experiments and generated the sequencing data; D.P.R. designed and conducted the bioinformatics analysis; M.J.M., D.P.R., and M.M.D. analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Genome sequence data have been deposited in GenBank under BioProject identifier PRJNA308843.
Extended data figures and tables
Extended Data Figure 1 Genetic system and experimental protocol for evolution of sexual populations.
Genotypes of the two haploid mating types are indicated at bottom, with selectable markers that are expressed in each strain indicated in colour. Steps in our experimental protocols involving these markers are indicated in the corresponding colour. STE5pr is a haploid-specific promoter and STE2pr and STE3pr are a- and α-specific promoters respectively, so haploid a cells express URA3 and HIS3, while haploid α cells express URA3 and LEU2. The drug resistance markers KANMX and HPHB, tightly linked to the a and α mating loci respectively, are constitutively expressed. URA3 is counterselectable; it is not expressed in diploids, rendering them resistant to 5-FOA.
Extended Data Figure 2 Adaptation to 17 °C and sporulation conditions.
a, b, Relative fitness of evolved asexual (blue) and sexual (orange) populations over four days in 17 °C (a) and sporulation conditions (b). Fitness changes are reported averaged over a complete experimental cycle (90 generations; mean of three replicate fitness assays, error bars ± s.e.m.). Mean fitness differences between asexual and sexual evolved strains are not significant in either the 17 °C (two-sided t-test, P = 0.5) or sporulation (two-sided t-test, P = 0.8) treatment.
Extended Data Figure 3 Adaptation in mixed and non-mixed asexual populations.
Fitness increases after 990 generations of evolution in mixed (blue) and non-mixed (pink) alternative asexual control populations (mean of four replicate fitness measurements, error bars ± s.e.m.). Each non-mixed line was maintained independently. Subpopulations from mixed populations were mixed in pairs every 90 generations; each pair is indicated by a corresponding light and dark circle.
Extended Data Figure 4 Read-depth variation analysis of sequenced clones.
Denoised, normalized coverage in 100-bp windows along the genome (Methods). Each panel represents a clone isolated from one of four independent populations. Pairs of clones from the same population are adjacent and indicated by the population label on the left. Regions containing putative amplifications and deletions (Extended Data Table 4) are highlighted in orange.
Supplementary information
Supplementary Data 1
This tab-delimited text file contains the identity, frequency trajectory, and fitness data for all mutations in this study. (TXT 322 kb)
Supplementary Data 2
This excel file contains all primers and oligonucleotides used in this study. (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
McDonald, M., Rice, D. & Desai, M. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531, 233–236 (2016). https://doi.org/10.1038/nature17143
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17143
This article is cited by
-
Bacteria-phage coevolution with a seed bank
The ISME Journal (2023)
-
Embracing Complexity: Yeast Evolution Experiments Featuring Standing Genetic Variation
Journal of Molecular Evolution (2023)
-
Crossover patterning in plants
Plant Reproduction (2023)
-
Evolution of sex determination in crustaceans
Marine Life Science & Technology (2023)
-
Improved biomass production of a microalga through adaptative laboratory evolution to a high light environment
Journal of Applied Phycology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.