Abstract
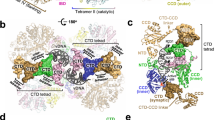
Retroviral integrase catalyses the integration of viral DNA into host target DNA, which is an essential step in the life cycle of all retroviruses1. Previous structural characterization of integrase–viral DNA complexes, or intasomes, from the spumavirus prototype foamy virus revealed a functional integrase tetramer2,3,4,5, and it is generally believed that intasomes derived from other retroviral genera use tetrameric integrase6,7,8,9. However, the intasomes of orthoretroviruses, which include all known pathogenic species, have not been characterized structurally. Here, using single-particle cryo-electron microscopy and X-ray crystallography, we determine an unexpected octameric integrase architecture for the intasome of the betaretrovirus mouse mammary tumour virus. The structure is composed of two core integrase dimers, which interact with the viral DNA ends and structurally mimic the integrase tetramer of prototype foamy virus, and two flanking integrase dimers that engage the core structure via their integrase carboxy-terminal domains. Contrary to the belief that tetrameric integrase components are sufficient to catalyse integration, the flanking integrase dimers were necessary for mouse mammary tumour virus integrase activity. The integrase octamer solves a conundrum for betaretroviruses as well as alpharetroviruses by providing critical carboxy-terminal domains to the intasome core that cannot be provided in cis because of evolutionarily restrictive catalytic core domain–carboxy-terminal domain linker regions. The octameric architecture of the intasome of mouse mammary tumour virus provides new insight into the structural basis of retroviral DNA integration.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Coordinates of cryo-EM density maps for the full and core intasome datasets have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-6440 and EMD-6441, respectively. X-ray diffraction data and the resulting INCCD, INNTD–CCD and INCTD structures have been deposited in the Protein Data Bank (PDB) under accession numbers 5CZ1, 5CZ2 and 5D7U, respectively. The core intasome structure has been deposited in the Protein Data Bank under accession number 3JCA.
References
Craigie, R. & Bushman, F. D. HIV DNA integration. Cold Spring Harb. Perspect. Med. 2, a006890 (2012)
Hare, S., Gupta, S. S., Valkov, E., Engelman, A. & Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 (2010)
Maertens, G. N., Hare, S. & Cherepanov, P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468, 326–329 (2010)
Hare, S., Maertens, G. N. & Cherepanov, P. 3′-Processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 31, 3020–3028 (2012)
Maskell, D. P. et al. Structural basis for retroviral integration into nucleosomes. Nature 523, 366–369 (2015)
Yang, Z. N., Mueser, T. C., Bushman, F. D. & Hyde, C. C. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 296, 535–548 (2000)
Wang, J. Y., Ling, H., Yang, W. & Craigie, R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20, 7333–7343 (2001)
Bao, K. K. et al. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 278, 1323–1327 (2003)
Li, M., Mizuuchi, M., Burke, T. R., Jr & Craigie, R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25, 1295–1304 (2006)
Hare, S. et al. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 5, e1000259 (2009)
Majors, J. E. & Varmus, H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature 289, 253–258 (1981)
Roth, M. J., Schwartzberg, P. L. & Goff, S. P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 58, 47–54 (1989)
Craigie, R., Fujiwara, T. & Bushman, F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62, 829–837 (1990)
Ellison, V. & Brown, P. O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc. Natl Acad. Sci. USA 91, 7316–7320 (1994)
DiMaio, F. et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nature Methods 12, 361–365 (2015)
Kudryashev, M. et al. Structure of the type VI secretion system contractile sheath. Cell 160, 952–962 (2015)
Wang, R. Y. et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature Methods 12, 335–338 (2015)
van Gent, D. C., Vink, C., Groeneger, A. A. & Plasterk, R. H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 12, 3261–3267 (1993)
Engelman, A., Bushman, F. D. & Craigie, R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12, 3269–3275 (1993)
Yang, F. & Roth, M. J. Assembly and catalysis of concerted two-end integration events by Moloney murine leukemia virus integrase. J. Virol. 75, 9561–9570 (2001)
Diamond, T. L. & Bushman, F. D. Division of labor within human immunodeficiency virus integrase complexes: determinants of catalysis and target DNA capture. J. Virol. 79, 15376–15387 (2005)
Baker, T. A., Mizuuchi, M., Savilahti, H. & Mizuuchi, K. Division of labor among monomers within the Mu transposase tetramer. Cell 74, 723–733 (1993)
Jenkins, T. M., Esposito, D., Engelman, A. & Craigie, R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 16, 6849–6859 (1997)
Yin, Z., Shi, K., Banerjee, S., Grandgenett, D. P. & Aihara, H. Crystal structure of the Rous sarcoma virus intasome. Nature http://dx.doi.org/10.1038/nature16950 (this issue)
Faure, A. et al. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 33, 977–986 (2005)
Bera, S., Pandey, K. K., Vora, A. C. & Grandgenett, D. P. Molecular interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J. Mol. Biol. 389, 183–198 (2009)
Lee, S. P., Xiao, J., Knutson, J. R., Lewis, M. S. & Han, M. K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 36, 173–180 (1997)
Heuer, T. S. & Brown, P. O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry 37, 6667–6678 (1998)
Serrao, E., Ballandras-Colas, A., Cherepanov, P., Maertens, G. N. & Engelman, A. N. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology 12, 39 (2015)
Engelman, A. & Craigie, R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66, 6361–6369 (1992)
Ballandras-Colas, A., Naraharisetty, H., Li, X., Serrao, E. & Engelman, A. Biochemical characterization of novel retroviral integrase proteins. PLoS ONE 8, e76638 (2013)
Cherepanov, P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35, 113–124 (2007)
Demeler, B. et al. UltraScan-III version 2.2: a comprehensive data analysis software package for analytical ultracentrifugation experiments http://www.ultrascan3.uthscsa.edu/ (2014)
Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds Harding, S. E., Rowe, A. J. & Horton, J. C. ) 90–125 (Royal Society of Chemistry, 1992)
Demeler, B. in Current Protocols in Protein Science (eds Coligan, J. E. et al.) Ch. 7, Unit 7.13, 7.13.1–7.13.24 (Wiley, 2010)
Brookes, E., Cao, W. & Demeler, B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 39, 405–414 (2010)
Schuck, P. & Demeler, B. Direct sedimentation analysis of interference optical data in analytical ultracentrifugation. Biophys. J. 76, 2288–2296 (1999)
Brookes, E. H. & Demeler, B. in Proceedings of the 9th Annual Conference on Genetic and Evolutionary Computation 361–368 (Association for Computing Machinery, 2007)
Demeler, B. & Brookes, E. Monte Carlo analysis of sedimentation experiments. Colloid Polym. Sci. 286, 129–137 (2008)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Bujacz, G. et al. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 253, 333–346 (1995)
Chen, J. C.-H. et al. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl Acad. Sci. USA 97, 8233–8238 (2000)
Morris, R. J., Perrakis, A. & Lamzin, V. S. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013)
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015)
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009)
Sorzano, C. O. et al. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 171, 197–206 (2010)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Lyumkis, D., Vinterbo, S., Potter, C. S. & Carragher, B. Optimod – an automated approach for constructing and optimizing initial models for single-particle electron microscopy. J. Struct. Biol. 184, 417–426 (2013)
Grigorieff, N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 (2007)
Lyumkis, D., Brilot, A. F., Theobald, D. L. & Grigorieff, N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388 (2013)
DiMaio, F., Zhang, J., Chiu, W. & Baker, D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 22, 865–868 (2013)
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014)
Acknowledgements
We acknowledge support from US National Institutes of Health (NIH) grants R01 AI070042 (to A.N.E.), NIH P50 GM103368 and the Leona M. and Harry B. Helmsley Charitable Trust grant number 2012-PG-MED002 (to D.L., both funding sources provided equal support), NIH P50 GM082251 (to P.C.), NIH P30 AI060354 (Harvard University Center for AIDS Research), and US National Science Foundation grants NSF-ACI-1339649 and TG-MCB070039 (to B.D.). B.D. acknowledges support from San Antonio Cancer Institute grant CA054174 for the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio. Molecular graphics and analyses were performed with the USCF Chimera package (supported by NIH P41 GM103331). CryoEM data collection was in part facilitated by the National Resource for Automated Molecular Microscopy (9 P41 GM103310). We thank B. Anderson and J.-C. Ducom at The Scripps Research Institute for help with EM data collection and network infrastructure, J. Fitzpatrick and F. Dwyer for computational support at The Salk Institute, V. Pye for help with X-ray structure refinement and the staff of BM14 (European Synchrotron Radiation Facility, Grenoble, France) and I03 (Diamond Light Source, Oxfordshire, UK) beamlines for assistance with data collection.
Author information
Authors and Affiliations
Contributions
A.B.-C. and A.N.E. discovered how to assemble MMTV intasomes; A.B.-C. and T.G.D. expressed and purified MMTV IN proteins for biochemical analysis; A.B.-C. assembled intasomes, characterized their biochemistry, supplied them for cryo-EM and centrifugation analyses, and performed IN activity assays; M.B. and D.L. performed EM work, collected cryo-EM data and determined the structure; D.L. modelled the intasome structure; B.D. collected and analysed the sedimentation velocity data; N.J.C. and P.C. expressed and purified INCCD, INNTD–CCD and INCTD/212–266 constructs, established crystallization conditions and determined these structures.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Cryo-EM data and refinement.
a, Representative cryo-electron micrograph of MMTV intasomes, taken at 2.7 μm underfocus. b, Same as in a, marked to show selected particles. c, Two-dimensional class averages calculated using Relion56. d, Initial model from the class averages calculated using OptiMod57. e, Refined reconstruction from the full data set, with an Euler angle distribution plot showing the relative orientations of the particles.
Extended Data Figure 2 Cryo-EM resolution analysis of reconstructed intasome maps.
a, Fourier shell correlation curve corresponding to the refined map generated from the full intasome data set. b, Fourier shell correlation curve corresponding to the refined map generated from the core intasome data set with the NTDs, CCDs and interdomain linker regions of the flanking IN dimers computationally subtracted. Average global resolutions in a and b are indicated. c, Refined map generated from the full data set (left) displayed side-by-side with the same map coloured for local resolution (right). d, Refined map generated from the core intasome data set (left) displayed side-by-side with the same map coloured for local resolution (right) using the colouring scheme in c. e, Rotational snapshots of segmented density of CCD1 with the fit of the refined model (see Extended Data Fig. 6) highlighting structural features evident at ~4–5 Å resolution. Partial separation of β-strands, which is typically evident at or beyond 4.5 Å resolution, is apparent.
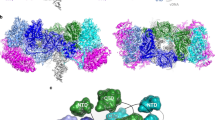
Extended Data Figure 3 Structural heterogeneity of the MMTV intasome.
a, Stable structural conformation of the MMTV intasome after three-dimensional classification of the data. Slices from the density map are displayed below. b, One of several conformations of MMTV intasome refinement after three-dimensional classification of the data. Slices from the density map are displayed below. Multiple fuzzy regions in the flanking INs are apparent in b, which are indicative of remaining heterogeneity within the data and/or continuous structural mobility of the region. c, Overlay of the two reconstructed maps, highlighting the extent of mobility within the flanking regions (brackets).
Extended Data Figure 4 MMTV IN domains and intasome sedimentation coefficient distribution.
a, Primary IN sequence alignment with boxes denoting canonical IN structural domains. The N-terminal extension domain occurs in spuma-, gamma- and epsilonretroviral IN proteins. Identical residues between MMTV, Rous sarcoma virus, HIV-1 and PFV INs are highlighted by red background; residues that are minimally conserved in three of the sequences are in red. PFV IN secondary structure elements are from PDB accession number 3L2Q; MMTV elements are from the INNTD–CCD and INCTD crystal structures described here (PDB accession numbers 5CZ2 and 5D7U, respectively). Symbols α, β, η, TT and TTT represent α-helix, β-strand, 310-helix, α-turn and β-turn, respectively. Figure generated with ESPript 3.0 (ref. 61). b, Monte Carlo analysis of sedimentation velocity data for the higher loading concentrations of vDNA (green), MMTV IN (blue) and intasome (red). A clear shift to a discrete species at 10.5 s is observed for the intasome, with minor IN and vDNA populations evident. Different centrifugation parameters for IN and vDNA versus intasomes (see Methods) probably attributed to the minor variations in sedimentation coefficient between major and minor IN and vDNA species. Measured sedimentation coefficients and calculated molar masses compared with theoretical molar masses are shown beneath the graph.
Extended Data Figure 5 MMTV IN domain crystal structures.
a, Stereo view of the final 2Fo − Fc density map of the INCCD crystal structure with blue mesh contoured at 1σ. Amino-acid side chains are readily evident at the 1.7 Å resolution. b, Stereo view of the final 2Fo − Fc density map of the 2.7 Å resolution INNTD–CCD crystal structure with blue mesh contoured at 1σ. The map is centred on the DDE catalytic triad (red sticks); green spheres, Mg2+ ions. c, Cartoon representation of the INCCD monomer (one of four in the crystallographic asymmetric unit) coloured in gold. Active site residues are shown as red sticks. d, Cartoon representation of the INNTD–CCD dimer structure (one of three in the asymmetric unit). The NTD and CCD are coloured green and gold, respectively. Red sticks, active site residues; grey and green spheres, Zn2+ and Mg2+ ions, respectively. e, Stereo view of the final 2Fo − Fc density map of the 1.5 Å resolution INCTD crystal structure, shown as a green mesh contoured at 1σ. f, Cartoon representation of one of the two CTD monomers present in the asymmetric unit.
Extended Data Figure 6 Molecular modelling of cryo-EM density.
a, Stereo views showing comparisons between the starting X-ray domain models and refined cryo-EM domain models for IN1 highlight relatively minor structural perturbations that are evident only in the most flexible regions of the intasome. b, Linker region snapshots. Atomic models were built de novo from the cryo-EM density for the indicated linkers in the top two panels (residues 45–54 connecting NTD1 and CCD1 and CCD–CTD residues 211–213). Linkers NTD2–CCD2, CCD5–CTD5 and CCD6–CTD6 were not modelled, but are shown as cryo-EM density (red) in the lower panels. c, Stereo view of the cryo-EM model for the MMTV intasome core region (Extended Data Fig. 2d), generated using Rosetta15,16,17. All domains were refined starting with the X-ray crystal structures (Extended Data Fig. 5). Specific linker regions were built de novo (continuous red lines) from the cryo-EM density, whereas lower-resolution linker regions (red dotted lines) were omitted from the model. d, Fourier shell correlation curve between the refined cryo-EM core intasome model and map, showing an average resolution of 4.8 Å. e, Comparison of two NTD–CCD conformations in the intasome highlights the NTD–CCD linker, which assumes a retracted state in the outer IN2 and IN4 monomers of core intasome dimers A and B, respectively, as well as in flanking IN dimers C and D (left). The linker extends in core IN molecules IN1 and IN3, which interact with the vDNA (right).
Extended Data Figure 7 Gel filtration profiles of INWT and IN mutant proteins.
Elution profiles of mass standards in kilodaltons as well as theoretical protein monomer (M) and dimer (D) positions are indicated.
Extended Data Figure 8 Comparisons of PFV and MMTV intasome structures.
a, Cartoon representations of the inner IN3 green subunits of the MMTV and PFV intasomes (Fig. 3a; vDNA strands are in grey). CCD–CTD linker regions are highlighted in orange, and dashed lines circle analogously positioned CTDs. Of note, this CTD in the MMTV structure is coloured differently because it originates from a separate IN molecule (IN8 from flanking dimer D). b, Lengths of NTD–CCD and CCD–CTD interdomain linker regions across retroviral IN proteins; ‘+’ indicates the presence of an N-terminal extension domain (NED). The multimeric state of IN in known intasome structures is indicated by bold type. c, The PFV intasome with bound tDNA (PDB accession number 3OS2; orange) was superimposed with the MMTV intasome (blue). The distance between overlaid active sites is in each case ~26 Å. d, Ninety-degree rotation of superimposed structures, with proteins omitted for clarity. Canonical B-form tDNA (magenta) was superimposed with PFV intasome tDNA. The positions of phosphodiester bonds staggered by 4 bp in the PFV crystal structure or by 6 bp in the modelled tDNA are indicated by spheres.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 – source data gels for Figures 1c and 4a. (PDF 423 kb)
Catalytic core domain in segmented electron density map
Catalytic core domain in segmented electron density map (MOV 12423 kb)
N-terminal domain in segmented electron density map
N-terminal domain in segmented electron density map (MOV 14435 kb)
C-terminal domain in segmented electron density map
C-terminal domain in segmented electron density map. (MOV 8560 kb)
Viral DNA in segmented electron density map
Viral DNA in segmented electron density map. (MOV 5632 kb)
MMTV intasome structure in electron density map with close up views of NTD1-CCD1, CCD1-CTD1 and CCD2-CTD2 linker regions
MMTV intasome structure in electron density map with close up views of NTD1-CCD1, CCD1-CTD1 and CCD2-CTD2 linker regions (MOV 24825 kb)
Source data
Rights and permissions
About this article
Cite this article
Ballandras-Colas, A., Brown, M., Cook, N. et al. Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature 530, 358–361 (2016). https://doi.org/10.1038/nature16955
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16955
This article is cited by
-
A proteomic screen of Ty1 integrase partners identifies the protein kinase CK2 as a regulator of Ty1 retrotransposition
Mobile DNA (2022)
-
Structure and function of retroviral integrase
Nature Reviews Microbiology (2022)
-
INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication
Nature Communications (2021)
-
Influence of the amino-terminal sequence on the structure and function of HIV integrase
Retrovirology (2020)
-
Structural basis of host protein hijacking in human T-cell leukemia virus integration
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.