Abstract
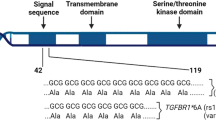
Variant rs351855-G/A is a commonly occurring single-nucleotide polymorphism of coding regions in exon 9 of the fibroblast growth factor receptor FGFR4 (CD334) gene (c.1162G>A). It results in an amino-acid change at codon 388 from glycine to arginine (p.Gly388Arg) in the transmembrane domain of the receptor. Despite compelling genetic evidence for the association of this common variant with cancers of the bone1, breast2, colon3, prostate4,5, skin6, lung7,8, head and neck9, as well as soft-tissue sarcomas and non-Hodgkin lymphoma, the underlying biological mechanism has remained elusive. Here we show that substitution of the conserved glycine 388 residue to a charged arginine residue alters the transmembrane spanning segment and exposes a membrane-proximal cytoplasmic signal transducer and activator of transcription 3 (STAT3) binding site Y390-(P)XXQ393. We demonstrate that such membrane-proximal STAT3 binding motifs in the germline of type I membrane receptors enhance STAT3 tyrosine phosphorylation by recruiting STAT3 proteins to the inner cell membrane. Remarkably, such germline variants frequently co-localize with somatic mutations in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. Using Fgfr4 single nucleotide polymorphism knock-in mice and transgenic mouse models for breast and lung cancers, we validate the enhanced STAT3 signalling induced by the FGFR4 Arg388-variant in vivo. Thus, our findings elucidate the molecular mechanism behind the genetic association of rs351855 with accelerated cancer progression and suggest that germline variants of cell-surface molecules that recruit STAT3 to the inner cell membrane are a significant risk for cancer prognosis and disease progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
29 January 2016
The ProteomeXchange Consortium accession code was corrected on 29 January 2016
References
Morimoto, Y. et al. Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer 98, 2245–2250 (2003)
Frullanti, E. et al. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur. J. Cancer Prevention 20, 340–347 (2011)
Heinzle, C. et al. Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 72, 5767–5777 (2012)
Xu, W. et al. FGFR4 transmembrane domain polymorphism and cancer risk: a meta-analysis including 8555 subjects. Eur. J. Cancer 46, 3332–3338 (2010)
Xu, B. et al. FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer 11, 84 (2011)
Streit, S., Mestel, D. S., Schmidt, M., Ullrich, A. & Berking, C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. Br. J. Cancer 94, 1879–1886 (2006)
Spinola, M. et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J. Clin. Oncol. 23, 7307–7311 (2005)
Falvella, F. S. et al. FGFR4 Gly388Arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int. J. Cancer 124, 2880–2885, (2009)
Streit, S. et al. Involvement of the FGFR4 Arg388 allele in head and neck squamous cell carcinoma. Int. J. Cancer 111, 213–217 (2004)
Bange, J. et al. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 62, 840–847 (2002)
Abecasis, G. R. et al.; 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Sharpe, H. J., Stevens, T. J. & Munro, S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142, 158–169 (2010)
Buss, J. E., Der, C. J. & Solski, P. A. The six amino-terminal amino acids of p60src are sufficient to cause myristylation of p21v-ras. Mol. Cell. Biol. 8, 3960–3963 (1988)
Shao, H. et al. Structural requirements for signal transducer and activator of transcription 3 binding to phosphotyrosine ligands containing the YXXQ motif. J. Biol. Chem. 279, 18967–18973 (2004)
Cunningham, F. et al. Ensembl 2015. Nucleic Acids Res . 43, D662–D669 (2015)
Hancock, J. F., Cadwallader, K., Paterson, H. & Marshall, C. J. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 10, 4033–4039 (1991)
Rotin, D. et al. SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C gamma. EMBO J. 11, 559–567 (1992)
Sandgren, E. P., et al. Inhibition of mammary gland involution is associated with transforming growth factor α but not c-myc-induced tumorigenesis in transgenic mice. Cancer Res . 55, 3915–3927 (1995)
Parker, R. A. & Bermann, N. G. Sample size: more than calculations. Am. Stat. 57, 166–170 (2003)
Vizcaíno, J. A., et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnol. 32, 223–226 (2014)
Acknowledgements
We thank the NHLBI GO Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010). We also thank N. Nagaraj for the help with quantitative proteomics and T. W. J. Gadella for providing us pmTurquoise2-N1 and C1 plasmids. We acknowledge T. Mayr for helping us by providing Fgfr4 knock-in mice for preliminary experiments, and R. Hornberger for technical assistance. We thank M. Sibilia for providing the Egfr knockout and wild-type counterpart MEFs. R. Abraham provided FGFR4 extracellular domain-specific anti-human mAb, clone 4FA6#11. We also acknowledge H. Brandstetter, S. Uebel and M. Spitaler for their assistance and services.
Author information
Authors and Affiliations
Contributions
A.U. conceptualized and planned the project. A.U. and V.K.U. designed the study. A.U. and V.K.U. coordinated the experimental work and wrote the manuscript with input from co-authors. V.K.U. investigated, analysed and interpreted the results. V.K.U. planned and performed in vitro and in vivo experiments with assistance from B.S.; normal healthy knock-in mice were generated and analysed by V.K.U.; the knock-in mouse model for lung cancer Fgfr4G/G;SPC-CrafBxB and Fgfr4A/A;SPC-CrafBxB was generated and analysed by V.K.U.; U.R.R. provided SPC-CrafBxB breeder pairs for this study. The knock-in mouse model for breast cancer Fgfr4G/G;WAP-Tgfα and Fgfr4A/A;WAP-Tgfα was generated by B.S. and analysed by V.K.U.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Effect of the rs351855 SNP in the knock-in MEFs.
a, Human FGFR4 gene structure depicting the single nucleotide change (rs351855) from guanine to adenine in exon 9 (red arrow). The rs351855-G (ancestral) allele is conserved across mammals, whereas the rs351855-A allele 388 occurs in approximately 30% of humans (according to the 1000 Genomes Project data set). Hydrophobic aliphatic 388Ile/Val evolved to charged 388Arg. b, Histogram showing the frequencies of the rs351855-G/A (p.Gly388Arg) allele in humans, including Africans (AFR), Asians (ASN), Europeans (EUR) and Americans (AMR), according to the data obtained from the 1000 Genomes Project (released 14 October 2013). c–e, SNP sequence (c), transcript levels (d) and immunoblot analysis (e) for FGFR4, pERK1/2, ERK1/2 and tubulin, and, f, intracellular flow cytometry staining for FGFR4 expression in Fgfr4G/G and Fgfr4A/A MEFs. g, Relative levels of ADP and ATP in the Fgfr4A/A, Fgfr4G/A and Fgfr4G/G MEFs (means ± s.d., n = 10, **P < 0.01, ****P < 0.0001, two-tailed unpaired t-test, 95% confidence level). h, Intracellular free calcium levels in Fgfr4A/A and Fgfr4G/G MEFs (mean ± s.d., n = 5, ****P < 0.0001). i, Comparison of the total proteome of Fgfr4G/G and Fgfr4A/A MEFs by quantitative mass spectrometry (see Methods). Cell-cycle proteins, blue; DNA metabolism proteins, pink.
Extended Data Figure 2 Effect of rs351855 SNP expression on cell proliferation.
a, Principal component analysis of the total proteome of quadruplicate samples of Fgfr4G/G and Fgfr4A/A MEFs. b, A line plot of the quantitative mass spectrometry data shows the differentially regulated proteins belonging to four gene ontology categories, including cell cycle, DNA metabolism, cell adhesion and cell junction (cut off log2(fold change) ± 0.5, Benjamini–Hochberg-corrected P < 0.05). c, Quantification of proliferation in Fgfr4A/A and Fgfr4G/G MEFs (see Methods) (mean ± s.d., n = 5, *P < 0.05, ****P < 0.0001, two-tailed unpaired t-test with a 95% confidence level). d, The proliferation of immortalized MEFs derived from Fgfr4G/G and Fgfr4A/A mice that were stably transduced with retroviruses encoding human oncogenes, including KRAS-G12V, BRAF-V600E, CRAF-BxB, EGFR-WT, EGFR-L858R, EGFR-L858R+T790M and EGFR-DEL1 (means ± s.d., n = 5, using two-way ANOVA and Sidak’s multiple comparisons test).
Extended Data Figure 3 Effect of the rs351855 SNP on the transmembrane segment of FGFR4.
a, The surface expression of FGFR4 was detected using a homemade mouse anti-FGFR4 mAb that detects the extracellular domain of FGFR4 by flow cytometry staining. Istotype control: red; FGFR4 staining: turquoise blue. Rs351855 SNP genotyping data. b, Quantification of relative surface expression levels of FGFR4. c, FGFR4 mRNA expression. HPRT1 as internal standard. Black: FGFR4G/G; red: FGFR4A/A (means ± s.d., n = 3). d, Immunoblot analyses for total FGFR4 and pSTAT3 (Y705) expression in human lung cancer cell lines. Black: FGFR4G/G; red: FGFR4A/A. e, Co-localization analyses of FGFR4 and endoplasmic reticulum chaperone BiP/GRP78 proteins. FGFR4: green; BiP/GRP78: red; nucleus: blue (n = 12, Fgfr4G/G cells and n = 15, Fgfr4A/A cells). f, Co-localization rate (means ± s.d., n = 12–15, ***P < 0.001). g, Relative abundance of isoleucine (top), glycine (middle) and arginine (bottom) along the TMDs in plants (left column), fungi (middle column) and vertebrates (right column). The position relative to the cytosolic edge (arrow) of the TMD is on the horizontal axes, and the residue abundance is on the vertical axes. Graphical plots generated using an algorithm available at http://www.tmdsonline.org. h, Prediction of putative transmembrane segment in FGFR4 p.388G (black) and risk-variant FGFR4 p.388R (red) variants. Data obtained using algorithm at http://split4.pmfst.hr/split/4/.
Extended Data Figure 4 Identification of germline variants in CD molecules generating membrane-proximal STAT3 binding site.
a, Graphical representation of the transmembrane sequence alignment of all human single-pass type I membrane proteins. Consensus sequence logo depicts stacks of amino-acid symbols with symbol height indicating the relative frequency of the amino acid in that position. Tyrosine (Y): green; glutamine (Q): magenta. b, Germline variations (superscript) in human CD molecules that generate a membrane-proximal STAT3 binding motif. Transmembrane domain: grey; missense mutation: red; YXXQ motifs: underlined. c, SNP name, protein allele, somatic status and the associated cancers for variants indicated by superscript numbers. d, STAT3-dependent promoter activity in immortalized Fgfr4G/G and Fgfr4A/A MEFs stably expressing the indicated human oncogenes (means ± s.d., n = 6, two-tailed non-parametric t-test, *P < 0.05, **P < 0.01 and ***P < 0.001). e, Immunoblot analyses for expression of pSTAT3 (Y705) in immortalized Fgfr4G/G, Fgfr4G/A and Fgfr4A/A MEFs stably expressing the indicated human oncogenes. f, Immunoblot analyses for expression of FGFR4 after knockdown of STAT3 in Fgfr4G/G, Fgfr4G/A and Fgfr4A/A MEFs. g, Proliferation analyses of Fgfr4G/G, Fgfr4G/A and Fgfr4A/A MEFs after STAT3 knockdown (means ± s.e.m., n = 5, *P < 0.05, **P < 0.01 and ***P < 0.001, two-way ANOVA and Tukey’s multiple comparison test). Note the increased sensitivity of Fgfr4A/A MEFs to the suppression of proliferation upon STAT3 knockdown.
Extended Data Figure 5 Effect of rs351855 SNP on sensitivity to growth inhibitors.
a, Flow cytometry analyses of co-cultivated Fgfr4G/G (GFP+) and Fgfr4A/A (RFP+) MEFs treated with the indicated growth-inhibiting drugs. Scatter plot depicts total cells (top) and live cells (bottom). b, Quantification of the relative proportion of live cells remaining after treatment (means ± s.d., n = 10, two-tailed non-parametric t-test, ***P < 0.001). c, Quantification of total Fgfr4G/G (GFP+) and Fgfr4A/A (RFP+) MEF live cells remaining in the co-culture, indicative of the success of growth inhibition. d, Expression of pSTAT3 (Y705) and total STAT3 expression in the co-cultivated Fgfr4G/G (GFP+) and Fgfr4A/A (RFP+) MEFs. e, Knockdown of STAT3 and EGFR expression in co-cultivated MEFs. f, Flow cytometry analyses of co-cultivated Fgfr4G/G (GFP+) and Fgfr4A/A (RFP+) MEFs after siRNA transfection. Dot-plot images are representative of three independent experiments. g, Relative quantification of Fgfr4G/G (GFP+) and Fgfr4A/A (RFP+) MEFs (means ± s.d., n = 6, two-way ANOVA and Sidak’s multiple comparison test, ****P < 0.0001).
Extended Data Figure 6 Identification of tyrosine-390 phosphorylation in the FGFR4 Arg388 variant.
a, Co-localization analyses for STAT3–CFP and FGFR4 Gly388Δ–YFP and FGFR4 Arg388Δ–YFP variants in HEK293T transfectants (means ± s.e.m., n = 26, ****P < 0.0001, one-tailed unpaired t-test with Welch’s correction). b, Immunoblot detection of phosphorylated tyrosines in purified FGFR4 Gly388Δ–YFP–His and FGFR4 Arg388Δ–YFP–His recombinant proteins. c, Mass spectrometry identification of Y-390 in FGFR4 Arg388Δ–YFP variant. Selected peptide-spectrum matches and the ion table displaying the calculated mass of the possible fragment ions are shown. N-terminal ions: blue; C-terminal ions: red.
Extended Data Figure 7 Direct interaction of STAT3 with the membrane-proximal YXXQ motifs.
a, Representative CFP, YFP and FRET ratio images of HEK293T transfectants. Co-transfection with FGFR4 Arg388Δ–YFP and STAT3–turquoise-N1 (membrane-targeted STAT3) served as a reference control for the FRET calculations. Data shown are representative of five independent FRET imaging experiments. b, Quantification of FRET efficiencies calculated for the selected cell membrane as region of interest (ROI) (see Methods). Data shown are representative of five independent FRET imaging experiments (means ± s.e.m., n = 12 cells; ***P < 0.001, ****P < 0.0001). c, Quantification of STAT3-dependent promoter activity by FGFR4 Arg388 variant lacking intracellular kinase domain in HEK293T transfectants (means ± s.e.m., n = 6, **P < 0.01, two-tailed unpaired t-test, 95% confidence level). d, Assessment of intracellular and surface levels of the transfected transmembrane peptides by flow cytometry (see Methods). e, STAT3-dependent promoter activity in HEK293T cells transfected with the indicated peptides (means ± s.e.m., n = 3, ***P < 0.001). f, Immunoblot analyses for pSTAT3 (Y705) expression in peptide transfectants.
Extended Data Figure 8 Localization and phosphorylation of membrane-targeted STAT3.
a, Representative confocal images of HEK293T transfectants. First column: FRET ratiometric images; second column: STAT3 localization; third column: pSTAT3 (Y705) localization; fourth column: overlay of pSTAT3 (Y705) and differential interference contrast images; fifth column enlarged images from selected (yellow rectangle) region. Magnification ×63. Images are representative of ten acquisitions. b, Immunoblot analyses of pSTAT3 (Y705) expression in HEK293T transfectants. The YXXQ motif was introduced at the juxtamembrane region (L414Y), tyrosine kinase domain (V550Q) and cytoplasmic tail terminus (L757Q) in the wild-type FGFR4 p.G388 variant. As a control, the YXXQ motif in risk variant FGFR4 p.R388 was destroyed by a mutation of Y390A. c, Quantification of colony formation assay results (see Methods). Shown are the representative results of three independent assays (means ± s.e.m., n = 4 wells, *P < 0.05; ns, not significant).
Extended Data Figure 9 FGFR4 p.G388R-induced STAT3 activation is dependent on EGFR.
a–c, Immunoblot analyses for pSTAT3 (Y705) expression in Fgfr4G/G and Fgfr4A/A MEFs treated with EGFR inhibitor (a), JAK inhibitor (b) and 10 μM TGI-101348 (JAK inhibitor), 1 μM wortmannin and 50 μM LY24002 (PI3K inhibitor) (c). d, Immunoblot analyses for pSTAT3 (Y705) levels in Fgfr4G/G and Fgfr4A/A MEFs after EGFR knockdown. e, Immunoblot analyses for pSTAT3 (Y705) levels in Egfr+/+ and Egfr−/− MEFs transfected either with plasmids encoding the Fgfr4 Gly388Δ–YFP and Fgfr4 Arg388Δ–YFP variants or with the transmembrane peptides, namely peptide-tmGly388 (rs351855-G) and peptide-tmArg388 (rs351855-A). f, g, STAT3-dependent promoter activity in Egfr+/+ and Egfr−/− MEFs transfected with plasmids (f) and peptides (g). The results are representative of three independent experiments (means ± s.e.m., n = 3, ***P < 0.001 and **P < 0.01).
Extended Data Figure 10 Human germline variants affecting the membrane-proximal STAT3 binding site.
a, Summary of the results obtained from combined analyses of the Ensembl variation data set and NHLBI exome variant data set. Dim red boxes: common germline mutations; bright red boxes: rare germline mutations that give rise to a YXXQ motif. Scissored arrowheads: rare germline mutations that destroy the YXXQ motif either by a frame shift or deletion at or before the YXXQ motif in the DNA sequence. b, 1000 Genome allele frequencies of all the FGFR4 non-synonymous coding region germline variants. We used the data from the 1000 Genomes Project (released 14 October 2013). c, Graphical summary explaining the new molecular function acquired by the germline variant rs351855 in the FGFR4 transmembrane domain. Alteration of the FGFR4 transmembrane spanning segment, such that Y390 was now located in the cytoplasm and phosphorylated, thereby exposing the functional STAT3 binding site (Y390-[p]RGQ390). Consequently, membrane-proximal phosphate transfer reactions (yellow symbol) dependent on EGFR activity lead to STAT3 tyrosine-705 phosphorylation, resulting in enhanced STAT3 signalling in cells.
Supplementary information
Supplementary Information
This file contains uncropped blots. (PDF 12266 kb)
Supplementary Table 1
This table contains a list of the proteins altered in FgfrG/G and FgfrA/A MEFs, as determined by quantitative mass spectrometry analyses. Shown is the list of proteins that were significantly altered (p < 0.001) when four biological replicates from either genotype were compared. Data are available via ProteomeXchange with identifier PXD003135 (XLSX 2650 kb)
Supplementary Table 2
The results of the rs351855 SNP genotyping of human cancer cell lines are listed. Source data: http://figshare.com/s/3f3cb5d4705c11e593ba06ec4b8d1f61 (XLSX 283 kb)
Supplementary Table 3
Display of the germline variations in and proximal to the transmembrane segments of all human receptor tyrosine kinases (Sheet1). The respective SNP IDs and allelic information are listed in Sheet 2. (XLSX 14 kb)
Supplementary Table 4
Identification of the germline variants that create membrane proximal STAT3 binding sites. A summary of the results from the combined analyses of the Ensembl Human Variation Dataset, dbSNP dataset, NHLBI Exome Server Dataset and Cosmic Datasets is shown. Source data: http://figshare.com/s/3f3cb5d4705c11e593ba06ec4b8d1f61 (XLSX 20 kb)
Rights and permissions
About this article
Cite this article
Ulaganathan, V., Sperl, B., Rapp, U. et al. Germline variant FGFR4 p.G388R exposes a membrane-proximal STAT3 binding site. Nature 528, 570–574 (2015). https://doi.org/10.1038/nature16449
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16449
This article is cited by
-
Proteogenomic links to human metabolic diseases
Nature Metabolism (2023)
-
A rare case of highly differentiated follicular carcinoma in ovary with FGFR4 Gly388Arg polymorphism: a case report and literature review
Journal of Ovarian Research (2022)
-
Bilateral adrenal uptake of 123I MIBG scintigraphy with mild catecholamine elevation, the diagnostic dilemma, and its characteristics
Scientific Reports (2022)
-
RETRACTED ARTICLE: Genipin suppression of growth and metastasis in hepatocellular carcinoma through blocking activation of STAT-3
Journal of Experimental & Clinical Cancer Research (2020)
-
Recurrent secondary genomic alterations in desmoplastic small round cell tumors
BMC Medical Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.