Abstract
Circadian clocks are endogenous timers adjusting behaviour and physiology with the solar day1. Synchronized circadian clocks improve fitness2 and are crucial for our physical and mental well-being3. Visual and non-visual photoreceptors are responsible for synchronizing circadian clocks to light4,5, but clock-resetting is also achieved by alternating day and night temperatures with only 2–4 °C difference6,7,8. This temperature sensitivity is remarkable considering that the circadian clock period (~24 h) is largely independent of surrounding ambient temperatures1,8. Here we show that Drosophila Ionotropic Receptor 25a (IR25a) is required for behavioural synchronization to low-amplitude temperature cycles. This channel is expressed in sensory neurons of internal stretch receptors previously implicated in temperature synchronization of the circadian clock9. IR25a is required for temperature-synchronized clock protein oscillations in subsets of central clock neurons. Extracellular leg nerve recordings reveal temperature- and IR25a-dependent sensory responses, and IR25a misexpression confers temperature-dependent firing of heterologous neurons. We propose that IR25a is part of an input pathway to the circadian clock that detects small temperature differences. This pathway operates in the absence of known ‘hot’ and ‘cold’ sensors in the Drosophila antenna10,11, revealing the existence of novel periphery-to-brain temperature signalling channels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dunlap, J. C., Loros, J. J. & DeCoursey, P. J. Chronobiology: Biological Timekeeping (Sinauer Associates, 2004)
Ouyang, Y., Andersson, C. R., Kondo, T., Golden, S. S. & Johnson, C. H. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl Acad. Sci. USA 95, 8660–8664 (1998)
Bechtold, D. A., Gibbs, J. E. & Loudon, A. S. Circadian dysfunction in disease. Trends Pharmacol. Sci. 31, 191–198 (2010)
Helfrich-Förster, C., Winter, C., Hofbauer, A., Hall, J. C. & Stanewsky, R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30, 249–261 (2001)
Hughes, S., Jagannath, A., Hankins, M. W., Foster, R. G. & Peirson, S. N. Photic regulation of clock systems. Methods Enzymol. 552, 125–143 (2015)
Brown, S. A., Zumbrunn, G., Fleury-Olela, F., Preitner, N. & Schibler, U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 12, 1574–1583 (2002)
Wheeler, D. A., Hamblen-Coyle, M. J., Dushay, M. S. & Hall, J. C. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J. Biol. Rhythms 8, 67–94 (1993)
Maguire, S. E. & Sehgal, A. Heating and cooling the Drosophila melanogaster clock. Curr. Opin . Insect Sci. 7, 71–75 (2015)
Sehadova, H. et al. Temperature entrainment of Drosophila’s circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron 64, 251–266 (2009)
Florence, T. J. & Reiser, M. B. Neuroscience: hot on the trail of temperature processing. Nature 519, 296–297 (2015)
Gallio, M., Ofstad, T. A., Macpherson, L. J., Wang, J. W. & Zuker, C. S. The coding of temperature in the Drosophila brain. Cell 144, 614–624 (2011)
Glaser, F. T. & Stanewsky, R. Temperature synchronization of the Drosophila circadian clock. Curr. Biol. 15, 1352–1363 (2005)
Wolfgang, W., Simoni, A., Gentile, C. & Stanewsky, R. The Pyrexia transient receptor potential channel mediates circadian clock synchronization to low temperature cycles in Drosophila melanogaster . Proc. R. Soc. Lond. B 280, 20130959 (2013)
Rees, J. S. et al. In vivo analysis of proteomes and interactomes using parallel affinity capture (iPAC) coupled to mass spectrometry. Mol. Cell Proteomics 10, M110.002386 (2011)
Abuin, L. et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60 (2011)
Benton, R., Vannice, K. S., Gomez-Diaz, C. & Vosshall, L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila . Cell 136, 149–162 (2009)
Rytz, R., Croset, V. & Benton, R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897 (2013)
Petersen, L. K. & Stowers, R. S. A Gateway MultiSite recombination cloning toolkit. PLoS ONE 6, e24531 (2011)
Sayeed, O. & Benzer, S. Behavioral genetics of thermosensation and hygrosensation in Drosophila . Proc. Natl Acad. Sci. USA 93, 6079–6084 (1996)
Kaneko, H. et al. Circadian rhythm of temperature preference and its neural control in Drosophila . Curr. Biol. 22, 1851–1857 (2012)
Hamada, F. N. et al. An internal thermal sensor controlling temperature preference in Drosophila . Nature 454, 217–220 (2008)
Ni, L. et al. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila . Nature 500, 580–584 (2013)
Busza, A., Murad, A. & Emery, P. Interactions between circadian neurons control temperature synchronization of Drosophila behavior. J. Neurosci. 27, 10722–10733 (2007)
Venken, K. J. et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster . Nature Methods 6, 431–434 (2009)
Kaneko, M. & Hall, J. C. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422, 66–94 (2000)
Gummadova, J. O., Coutts, G. A. & Glossop, N. R. Analysis of the Drosophila Clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. J. Biol. Rhythms 24, 353–367 (2009)
Kim, J. et al. A TRPV family ion channel required for hearing in Drosophila . Nature 424, 81–84 (2003)
Park, J. H. & Hall, J. C. Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster . J. Biol. Rhythms 13, 219–228 (1998)
Liu, L. et al. Drosophila hygrosensation requires the TRP channels water witch and nanchung. Nature 450, 294–298 (2007)
Chung, Y. D., Zhu, J., Han, Y. & Kernan, M. J. nompA encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron 29, 415–428 (2001)
Ruben, M., Drapeau, M. D., Mizrak, D. & Blau, J. A mechanism for circadian control of pacemaker neuron excitability. J. Biol. Rhythms 27, 353–364 (2012)
Konopka, R. J. & Benzer, S. Clock mutants of Drosophila melanogaster . Proc. Natl Acad. Sci. USA 68, 2112–2116 (1971)
Sweeney, S. T., Broadie, K., Keane, J., Niemann, H. & O’Kane, C. J. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14, 341–351 (1995)
Zhang, Y., Liu, Y., Bilodeau-Wentworth, D., Hardin, P. E. & Emery, P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr. Biol. 20, 600–605 (2010)
Saina, M. & Benton, R. Visualizing olfactory receptor expression and localization in Drosophila . Methods Mol. Biol. 1003, 211–228 (2013)
Yoshii, T., Todo, T., Wülbeck, C., Stanewsky, R. & Helfrich-Förster, C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J. Comp. Neurol. 508, 952–966 (2008)
Rush, B. L., Murad, A., Emery, P. & Giebultowicz, J. M. Ectopic CRYPTOCHROME renders TIM light sensitive in the Drosophila ovary. J. Biol. Rhythms 21, 272–278 (2006)
Gentile, C., Sehadova, H., Simoni, A., Chen, C & Stanewsky, R . Cryptochrome antagonizes synchronization of Drosophila’s circadian clock to temperature cycles. Curr. Biol. 23, 185–195 (2013)
Croset, V. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6, e1001064 (2010)
Levine, J. D., Funes, P., Dowse, H. B. & Hall, J. C. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3, 1 (2002)
Simoni, A. et al. A mechanosensory pathway to the Drosophila circadian clock. Science 343, 525–528 (2014)
Wilson, R. I. & Corey, D. P. The force be with you: a mechanoreceptor channel in proprioception and touch. Neuron 67, 349–351 (2010)
Stanewsky, R. et al. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J. Neurosci. 17, 676–696 (1997)
Acknowledgements
We thank P. Emery, J. Albert, J. Jepson, P. Garrity, and A. Samuel for discussions and sharing of unpublished results, J. Giebultowicz for anti-TIM antibodies, J. Albert and J. Jepson for fly stocks, C. Tardieu and R. Kavlie for help with qPCR, D. Carr for assistance with the temperature recording setup, and M. Ogueta-Gutierrez for help with figure preparations. The drawing for Fig. 4a was generated by Polygonal Tree (http://polygonaltree.co.uk/). This work was supported by BBSRC grants BB/H001204 to R.S., BB/J0-18589/-17221 to R.S. and J.J.L.H., and a CSC PhD fellowship to C.C. V.C. was supported by a Boehringer Ingelheim Foundation Fellowship. Research in R.B.’s laboratory was supported by European Research Council Starting Independent Researcher and Consolidator Grants (205202 and 615094). Mass spectrometry analysis was supported by Wellcome Trust grant 099135/Z/12/Z.
Author information
Authors and Affiliations
Contributions
C.C., E.B., R.S., J.J.L.H., R.B. and K.S.L. conceived, designed, and supervised the project. C.C., E.B., M.X., V.C., and J.S.R. performed experiments. C.C., E.B., and J.S.R. analysed data, and R.S. wrote the paper, with feedback from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 IR25a and Nocte physically interact in vivo and are expressed in femur and antennal ChO neurons.
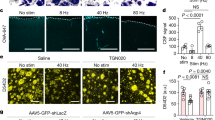
a, In vivo co-immunoprecipitation experiments using protein extracts from fly heads. Head lysates were immunoprecipitated using anti-Flag antibody. The immunoprecipitates were examined by western blotting using anti-Flag and anti-IR25a antibody. Input represents 30% of cell lysates used in the pull-down experiment. The genotypes of the flies used were: tim > IR25a: UAS-GFP/UAS-IR25a; tim-gal4:67/+. tim > IR25a+FLAG-NOCTE: UAS-FSNH/UAS-IR25a; tim-gal4:67/+. The bracket indicates that NOCTE–Flag runs as a double band on western blots. For uncropped gel images, see Supplementary Fig. 1. b, Overview of the antennal and femur ChO adapted from refs 13, 42. c, d, Labelling of the JO neurons by IR25a-gal4 (c) and F-gal4 (d) driven membrane bound mCD8–GFP and nuclear-localized DsRed expression. Note that IR25a is expressed in only a subset of JO neurons. e, f, Same flies as in c, d analysed for IR25a expression in the femoral ChO. Again, only subsets of the ChO neurons express IR25a. Arrows in c, e point to ChO neuron nuclei. g, Labelling of the ChO neurons in JO and femur by nocte-gal4-driven membrane-bound GFP and nuclear DsRed expression9. Scale bar, 20 μm.
Extended Data Figure 2 Spatial and quantitative IR25a mRNA and protein expression in CNS and PNS tissues and efficiency of RNAi-mediated knockdown.
a, Analysis of IR25a-gal4 and IR25a in the third segment of the antenna reveals expression in coeloconic sensilla16. Schematic adapted from15. b, c, Determination of IR25a and nocte mRNA levels in femur and retinal tissues by semiquantitative RT–PCR; rp49 was used as control. For uncropped gel data, see Supplementary Fig. 1. d, e, qPCR analysis of IR25a mRNA levels in whole heads (d), or dissected body parts (as indicated) (e) from flies of the genotypes indicated. Pan-neuronal elav-gal4 knockdown (d) decreased IR25a mRNA >75% or >90%, using one or two different RNAi lines combined, respectively. ****P < 0.0001, ***P < 0.001, *P < 0.05, one-way ANOVA followed by Bonferroni correction. f, IR25a is not expressed in the central brain and clock neurons. Left, IR25a immunolabelling of a Canton S brain reveals no signals. Middle, same brain labelled with anti-TIM reveals expression in clock neurons. Right, merge. Brains were dissected in LD at ZT20. Scale bar, 10 μm. g, IR25a-gal4 is not expressed in clock neurons and largely absent from the brain. Left, nuclear DsRed driven by IR25a-gal4. Second from left, anti-PDF staining showing LNv and their projections. Middle, anti-PER (diluted 1:5,000)43 showing all clock neurons. Second from right, merge, showing two IR25a-gal4 positive cell in the antennal lobe, not co-localized with any of the clock neurons. These cells were observed in 4/8 hemispheres and always on the same side of the brain. Right, magnified view of circled area in the merged image. Scale bar, 30 μm.
Extended Data Figure 3 IR25a is required for temperature synchronization to low-amplitude temperature cycles but not for high-amplitude temperature cycles.
a, Canton S, IR25a−/−, and nocte1 flies were exposed to LD at 20 °C for 5 days (left) or LL at 25 °C for 2 days (right), followed by exposure to a 12 h:12 h 20 °C:29 °C (left) or 16 °C:25 °C (right) temperature cycles in LL, which after 6–7 days was delayed or advanced by 6 h, respectively. Warmer temperature indicated by red and orange shading, respectively. b, Actograms and daily averages of Canton S and IR25a−/−, and IR25a−/− flies containing a genomic IR25a rescue construct (rescue) exposed to 18 °C:20 °C temperature cycles in LL (left) and 21 °C:23 °C temperature cycles in LL (right). Warm phase in actograms indicated by orange shading. Histogram colour coding as in Fig. 2. For quantification see Fig. 2e.
Extended Data Figure 4 IR25a is not required for temperature synchronization to high- but to low-amplitude temperature cycles and IR25a−/− flies show normal LD and DD behaviour.
a, Canton S and IR25a−/− flies were exposed to LL at 25 °C for 2–3 days, followed by exposure to high-amplitude 12 h:12 h temperature cycles in LL, which after 5–6 days were delayed by 6 h. Double plotted average actograms depicting the daily activity levels and environmental conditions during the entire experiment are shown. Actual temperatures are colour coded and indicated below the entrainment index calculations. Numbers (n) indicated in the bars. b, As in a but flies were initially kept in LD and DD for 2 days each (left) or LD (right), before being exposed to two phase delayed (left) or advanced (right) temperature cycles in DD at the temperatures indicated. n.s., not significant. c, Behaviour of IR25a−/− and rescue flies during DD and 25 °C:27 °C temperature cycles with 8 h delay and during DD and 21 °C:23 °C temperature cycles with a 8 h advance compared to the previous LD cycle (at 25 °C). Warm phase is indicated by orange shading. d, Canton S and IR25a−/− flies during LD and DD conditions at 25 °C (see Extended Data Table 2 for period calculations).
Extended Data Figure 5 Antennal IR25a expression is not necessary for synchronization of locomotor activity rhythms to temperature cycles.
Ablation of antennae as indicated. a, IR25a−/− and rescue flies were exposed to the same condition used in Fig. 2. Actograms and daily averages as described before. b, Quantification of behaviour as described in Fig. 2. The data of IR25a−/− with normal antennae was taken from Fig. 2a. n.s., not significant.
Extended Data Figure 6 Knocking down IR25a expression via RNA interference disrupts synchronization of locomotor activity rhythms to temperature cycles (25 °C:27 °C in LL).
a, b, Behaviour of flies with spatially restricted IR25a knockdown mediated by IR25a-gal4 (a), ChO specific F-gal4, and nompC-gal4 (b) driven IR25-RNAi expression, respectively. Control flies are UAS-dicer2/Y; IR25-gal4/+ (a), and UAS-dicer2/Y;+/+; F-gal4/+ or UAS-dicer2/Y;+/+; nompC-gal4/+ (b). Test flies carry the same transgenes, but in addition one or two copies of the IR25a-RNAi line indicated. Actograms and daily averages as described for Fig. 2a. c, Progeny of the respective UAS-IR25a-RNAi lines crossed to y w (left three columns) and flies from (a, b) and the other gal4 drivers indicated, were exposed to the same LL and temperature cycle conditions used in Fig. 2a. As controls, UAS-dicer, gal4 driver lines were crossed to y w and F1 males containing UAS-dicer/Y and the respective gal4/+ were tested. Numbers of analysed individuals (n) are indicated above each column. Entrainment was quantified as in Fig. 2c. ****P < 0.0001, one-way ANOVA followed by Bonferroni correction.
Extended Data Figure 7 Rescue of TIM oscillations in clock neurons during low-amplitude temperature cycles and normal TIM oscillations during LD and high-amplitude temperature cycles.
a, b, TIM levels in clock neurons during LL 25 °C:27 °C temperature cycles at the indicated time points (ZT) in the genotypes indicated. At least 8 brain hemispheres per time point were analysed for each genotype. Scale bars, 10 µm. Data in b are mean ± s.e.m. c, Quantification of TIM levels in clock neurons during LD (25 °C) in Canton S and IR25a−/− mutant brains. d, TIM oscillations in different clock-neuronal groups in IR25a−/− are restored in 25 °C:29 °C temperature cycles in LL. At least 8 brain hemispheres per time point were analysed for each genotype and condition. Error bars indicate s.e.m.
Extended Data Figure 8 Ectopic expression and heat responses of TRPA1 and IR8a in l-LNv clock neurons.
a, Whole-cell current clamp recordings of Pdf-gal4/UAS-TrpA1; Pdf-RFP (top trace, red) and Pdf-gal4/UAS-IR8a-RFP (bottom trace, black) brains exposed to a temperature ramp from 18 °C to 30 °C and back to 18 °C. Note the additional depolarization of the TrpA1 neuron at higher temperatures. b, Compared to control (Fig. 4), recordings from TrpA1-expressing neurons show a large increase in firing rate with temperature which the IR8a expressing neurons do not. c, d, In comparison to control neurons (data taken from Fig. 4) the membrane potential of TrpA1 expressing neurons is more positive at 30 °C (open bars) and the input resistance is also significantly reduced in TrpA1 at 18 °C. e, f, The firing rate at 18 °C is higher for IR8a neurons but only the Q10 of TrpA1 is different to control. Bars are means and whiskers s.e.m., n indicated in bars, *P < 0.05, ***P < 0.001, ANOVA followed by Tukey test.
Supplementary information
Supplementary Figure
This file contains gels for Extended Data Figures 1 and 2. (PDF 141 kb)
Rights and permissions
About this article
Cite this article
Chen, C., Buhl, E., Xu, M. et al. Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520 (2015). https://doi.org/10.1038/nature16148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16148
This article is cited by
-
Integration of photoperiodic and temperature cues by the circadian clock to regulate insect seasonal adaptations
Journal of Comparative Physiology A (2023)
-
A four-oscillator model of seasonally adapted morning and evening activities in Drosophila melanogaster
Journal of Comparative Physiology A (2023)
-
Ionotropic receptors in the turnip moth Agrotis segetum respond to repellent medium-chain fatty acids
BMC Biology (2022)
-
Animal behavior is central in shaping the realized diel light niche
Communications Biology (2022)
-
A natural timeless polymorphism allowing circadian clock synchronization in “white nights”
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.