Abstract
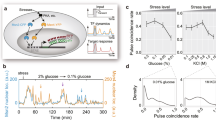
Studies of individual living cells have revealed that many transcription factors activate in dynamic, and often stochastic, pulses within the same cell. However, it has remained unclear whether cells might exploit the dynamic interaction of these pulses to control gene expression. Here, using quantitative single-cell time-lapse imaging of Saccharomyces cerevisiae, we show that the pulsatile transcription factors Msn2 and Mig1 combinatorially regulate their target genes through modulation of their relative pulse timing. The activator Msn2 and repressor Mig1 showed pulsed activation in either a temporally overlapping or non-overlapping manner during their transient response to different inputs, with only the non-overlapping dynamics efficiently activating target gene expression. Similarly, under constant environmental conditions, where Msn2 and Mig1 exhibit sporadic pulsing, glucose concentration modulated the temporal overlap between pulses of the two factors. Together, these results reveal a time-based mode of combinatorial gene regulation. Regulation through relative signal timing is common in engineering and neurobiology, and these results suggest that it could also function broadly within the signalling and regulatory systems of the cell.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits. (Taylor & Francis, 2006)
Gertz, J., Siggia, E. D. & Cohen, B. A. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457, 215–218 (2009)
Yosef, N. & Regev, A. Impulse control: temporal dynamics in gene transcription. Cell 144, 886–896 (2011)
Purvis, J. E. & Lahav, G. Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013)
Levine, J. H., Lin, Y. & Elowitz, M. B. Functional roles of pulsing in genetic circuits. Science 342, 1193–1200 (2013)
Lahav, G. et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nature Genet. 36, 147–150 (2004)
Nelson, D. E. et al. Oscillations in NF-κB signaling control the dynamics of gene expression. Science 306, 704–708 (2004)
Cai, L., Dalal, C. K. & Elowitz, M. B. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455, 485–490 (2008)
Locke, J. C., Young, J. W., Fontes, M., Hernández Jiménez, M. J. & Elowitz, M. B. Stochastic pulse regulation in bacterial stress response. Science 334, 366–369 (2011)
Hao, N. & O’Shea, E. K. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nature Struct. Mol. Biol. 19, 31–39 (2012)
Cohen-Saidon, C., Cohen, A. A., Sigal, A., Liron, Y. & Alon, U. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell 36, 885–893 (2009)
Young, J. W., Locke, J. C. & Elowitz, M. B. Rate of environmental change determines stress response specificity. Proc. Natl Acad. Sci. USA 110, 4140–4145 (2013)
Levine, J. H., Fontes, M. E., Dworkin, J. & Elowitz, M. B. Pulsed feedback defers cellular differentiation. PLoS Biol. 10, e1001252 (2012)
Garmendia-Torres, C., Goldbeter, A. & Jacquet, M. Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr. Biol. 17, 1044–1049 (2007)
Hao, N., Budnik, B. A., Gunawardena, J. & O’Shea, E. K. Tunable signal processing through modular control of transcription factor translocation. Science 339, 460–464 (2013)
Hansen, A. S. & O’Shea, E. K. Promoter decoding of transcription factor dynamics involves a trade-off between noise and control of gene expression. Mol. Syst. Biol. 9, 704 (2013)
Petrenko, N., Chereji, R. V., McClean, M. N., Morozov, A. V. & Broach, J. R. Noise and interlocking signaling pathways promote distinct transcription factor dynamics in response to different stresses. Mol. Biol. Cell 24, 2045–2057 (2013)
Kholodenko, B. N., Hancock, J. F. & Kolch, W. Signalling ballet in space and time. Nature Rev. Mol. Cell Biol. 11, 414–426 (2010)
Tay, S. et al. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 (2010)
Batchelor, E., Loewer, A., Mock, C. & Lahav, G. Stimulus-dependent dynamics of p53 in single cells. Mol. Syst. Biol. 7, 488 (2011)
Albeck, J. G., Mills, G. B. & Brugge, J. S. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261 (2013)
Yissachar, N. et al. Dynamic response diversity of NFAT isoforms in individual living cells. Mol. Cell 49, 322–330 (2013)
Kageyama, R., Ohtsuka, T., Shimojo, H. & Imayoshi, I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nature Neurosci. 11, 1247–1251 (2008)
Martínez-Pastor, M. T. et al. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235 (1996)
Schmitt, A. P. & McEntee, K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae . Proc. Natl Acad. Sci. USA 93, 5777–5782 (1996)
Boy-Marcotte, E., Perrot, M., Bussereau, F., Boucherie, H. & Jacquet, M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae . J. Bacteriol. 180, 1044–1052 (1998)
Estruch, F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24, 469–486 (2000)
Gasch, A. P. et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000)
Hasan, R. et al. The control of the yeast H2O2 response by the Msn2/4 transcription factors. Mol. Microbiol. 45, 233–241 (2002)
Morano, K. A., Grant, C. M. & Moye-Rowley, W. S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae . Genetics 190, 1157–1195 (2012)
Nehlin, J. O., Carlberg, M. & Ronne, H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10, 3373–3377 (1991)
Lutfiyya, L. L. & Johnston, M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 16, 4790–4797 (1996)
Carlson, M. Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207 (1999)
Teixeira, M. C. et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae . Nucleic Acids Res. 42, D161–D166 (2014)
Görner, W. et al. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12, 586–597 (1998)
De Vit, M. J., Waddle, J. A. & Johnston, M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8, 1603–1618 (1997)
Treitel, M. A., Kuchin, S. & Carlson, M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae . Mol. Cell. Biol. 18, 6273–6280 (1998)
Dalal, C. K., Cai, L., Lin, Y., Rahbar, K. & Elowitz, M. B. Pulsatile dynamics in the yeast proteome. Curr. Biol. 24, 2189–2194 (2014)
Shaner, N. C., Steinbach, P. A. & Tsien, R. Y. A guide to choosing fluorescent proteins. Nature Methods 2, 905–909 (2005)
Larson, D. R., Zenklusen, D., Wu, B., Chao, J. A. & Singer, R. H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–478 (2011)
Unnikrishnan, I., Miller, S., Meinke, M. & LaPorte, D. C. Multiple positive and negative elements involved in the regulation of expression of GSY1 in Saccharomyces cerevisiae . J. Biol. Chem. 278, 26450–26457 (2003)
Meister, M., Pine, J. & Baylor, D. A. Multi-neuronal signals from the retina: acquisition and analysis. J. Neurosci. Methods 51, 95–106 (1994)
De Wever, V., Reiter, W., Ballarini, A., Ammerer, G. & Brocard, C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 24, 4115–4123 (2005)
Ptashne, M. A Genetic Switch: Phage Lambda Revisited (Cold Spring Harbor Laboratory Press, 2004)
Bi, G. Q. & Poo, M. M. Synaptic modifications in cultured hippocampal neurons: dependence on spike timing, synaptic strength, and postsynaptic cell type. J. Neurosci. 18, 10464–10472 (1998)
Anderson, J. B., Aulin, T. & Sundberg, C.-E. Digital Phase Modulation (Springer, 1986)
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 31–34 (2007)
Sheff, M. A. & Thorn, K. S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae . Yeast 21, 661–670 (2004)
Goldstein, A. L. & McCusker, J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast 15, 1541–1553 (1999)
Lutfiyya, L. L. et al. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae . Genetics 150, 1377–1391 (1998)
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Unit 14.20 (2010)
Jaqaman, K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nature Methods 5, 695–702 (2008)
Teste, M. A., Duquenne, M., François, J. M. & Parrou, J. L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae . BMC Mol. Biol. 10, 99 (2009)
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 (2012)
Goecks, J., Nekrutenko, A. & Taylor, J. The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578 (2012)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Efron, B. The Jackknife, the Bootstrap, and Other Resampling Plans (Society for Industrial and Applied Mathematics, 1982)
Acknowledgements
We thank U. Alon, R. Corral, R. Deshaies, A. Eldar, J. Garcia-Ojalvo, R. Kishony, A. Moses, G. Seelig, P. Swain, and members of the Elowitz laboratory for comments and feedback on the manuscript. We also thank the core sequencing facility at Caltech for help on RNA-Seq. This work was supported by the NIH (R01 GM079771B, R01 GM086793A), the NSF (Award no. 1547056), DARPA (HR0011-05-1-0057), and by the Gordon and Betty Moore Foundation through Grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative. L.C. acknowledges the Ellison foundation for support.
Author information
Authors and Affiliations
Contributions
Y.L. and M.B.E. designed experiments. Y.L. performed experiments and analysed data with input from all authors. C.K.D. and L.C. initially observed the correlation between Msn2 and Mig1 dynamics and C.H.S. conducted preliminary analysis of target gene expression. M.B.E. supervised research. Y.L. and M.B.E. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Single-cell analysis of relative pulse timing modulation by stress identity during transient response.
a–c, Example traces for synthetic combinatorial (a), Msn2-specific (b), or Mig1-specific (c) promoters, in response to addition of 100 mM NaCl. Two cells are shown for each strain. For each cell, Msn2 and Mig1 localization traces (green and red) and the corresponding promoter response (blue) are shown on separate panels (top and bottom). Vertical dashed line indicates time of NaCl addition. d–f, Similar example traces for the response to addition of 2.5% ethanol. g, Average cross-correlation function of the transient Msn2 and Mig1 responses from t = 0–30 min after indicated stress. Cross-correlation between Msn2 and Mig1 is negative at time lag zero for both glucose reduction and NaCl stresses, but positive for ethanol stress. h, i, Averaged (left) and single-cell (right) nuclear localization traces of Msn2–mKO2 and Mig1–mCherry in response to 37 °C heat shock (h) or 0.25 mM H2O2 (i). j, k, Msn2 and Mig1 dynamics observed in Fig. 2b, c do not depend on the deletions introduced to the strain background. Averaged nuclear localization traces of Msn2–mKO2 and Mig1–mCherry in response to 100 mM NaCl (j) or 2.5% ethanol (k) for a control strain without msn4 mig2 deletions. Shading indicates 95% confidence intervals of the mean. l–n, Standard deviation representations of different sets of single-cell data (presented in main figures). The mean is indicated with a solid line, and ± 1 standard deviation ranges are indicated by shading. l, Nuclear localization responses of Msn2–mKO2 (green) and Mig1–mCherry (red) to downshift in glucose level (see Fig. 1b). m, n, Nuclear localizations and transcriptional responses to NaCl and ethanol. (see Fig. 2b, c).
Extended Data Figure 2 Additional data and analysis for transient stress responses.
a, Fold-change in expression in response to different stresses for synthetic combinatorial target gene for three genetic backgrounds: no deletion (MSN2 MIG1, data from Fig. 2f), msn2 deletion, and mig1 deletion. b, Similar plot for the endogenous target gene GSY1. Cells were treated with designated stress for 10 min and ≥ 3 biological replicates were averaged (error bar indicates s.e.m.). P value was obtained from two-tailed t-test. c, d, Averaged transcriptional responses of GSY1-24xPP7 in response to 100 mM NaCl (c) or 2.5% ethanol (d) for three genetic backgrounds: no deletion, mig1 deletion, and msn2 deletion. Averaged nuclear localization traces of Msn2–mKO2 and Mig1–mCherry for the ‘no deletion’ strain are shown on the top panels. e, Averaged nuclear localization traces of Msn2–mKO2 and Mig1–mCherry (top) and corresponding transcriptional responses for GSY1-24xPP7 in response to glucose downshift (from 0.2% to 0.1%). Shading in c–e indicates 95% confidence intervals of the mean. f–k, RNA-seq analysis (see Methods and Supplementary Discussion for more details). f, log2 fold-changes (LFC) in gene expression of 31 identified combinatorial targets (including GSY1; brown circle, indicated by green arrow) in response to NaCl (x axis) and ethanol (y axis) for wild-type background (that is, no deletion of either MSN2 or MIG1). g, The differences in LFC between wild-type and mig1 deletion for both NaCl (x axis) and ethanol (y axis). h, The differences in LFC between wild-type and msn2 deletion for both NaCl (x axis) and ethanol (y axis). i, The effect of Msn2 for each target was plotted against the corresponding number of Msn2 binding sites. j, Analogous plot for the effect of Mig1 binding sites. k, Correlation coefficients between the effect of Msn2 or Mig1 and the number of Msn2 or Mig1 binding motif, respectively. Error bars in f–h indicate standard deviations from two biological replicates. Error bars in k represent 95% confidence intervals from bootstrap.
Extended Data Figure 3 Example 3-colour single-cell traces under steady-state conditions, and schematic diagram of pulse-triggered averaging analysis.
a, b, Example 3-colour single-cell traces for synthetic (a) and natural (b) promoters under constant glucose (0.05%). Two cells are shown for each promoter. For each cell, nuclear localization traces are shown on the top and PP7–2 × GFP transcriptional output signal is shown on the bottom. c, Schematic illustration of pulse-triggered averaging analysis. Msn2 pulses were identified (green arrows) and sorted based on their relationship with the Mig1 signal within a 21 min time window (see Methods). Horizontal green and red lines underneath top time trace plot indicate width of identified Msn2 and Mig1 pulses, respectively. Msn2 pulses whose peaks overlap with Mig1 pulses were categorized as overlapping events (orange arrows) while the rest of Msn2 pulses were categorized as non-overlapping events (purple arrows). Overlapping and non-overlapping events were then averaged separately (bottom schematics).
Extended Data Figure 4 Pulse-triggered averaging analysis for control promoters and for delayed pulse timing events.
a, b, Plots analogous to those in Fig. 3d, e for additional synthetic and natural promoters. The GSY1 promoter was examined in strains with Msn2 or Mig1 zinc-finger deletions. For gene expression, areas under curves were analysed and presented in c, d. c, Relative pulse timing-dependent gene expression occurs for combinatorial promoters but not pure Msn2 or Mig1 target promoters. Bars represent integrated gene expression based on area under curve from Fig. 3d, e and a, b. d, Plot analogous to c for the natural GSY1 target gene. Binding of the transcription factors was abolished by mutations in zinc finger DNA-binding domains, indicated by crosses. e, Distributions of gene expression (estimated as integrated area under curve) per non-overlapping or overlapping event for both synthetic and natural combinatorial promoters (real data (solid) versus control data (dashed); top) and ratios between real and control data (bottom). Control data was measured from scrambled population of cells. For the real data, the distributions of non-overlapping and overlapping events are significantly different (by Kolmogorov–Smirnov test) with P values of 2.1 × 10−17 and 1.2 × 10−15 for synthetic and natural promoters, respectively. In contrast, for control data, they are not significantly different (P values: 0.4520 and 0.9888). For the calculation of ratios, averages of the non-overlapping and overlapping control data were used as control. f–i, Pulse-triggered averaging analysis of ‘delayed’ events in which an Msn2 pulse is followed by a Mig1 pulse (see Supplementary Discussion for details). f, Overlapping events were subdivided into delayed and non-delayed depending, as shown. Corresponding mean Msn2 and Mig1 signals as well as transcriptional responses were plotted for both synthetic and natural promoters. A similar classification was performed for non-overlapping events (g). Area under curve for f, g was plotted for direct comparison of gene expression between delayed and non-delayed pulse timing events (h, i). Shading and error bars indicate 95% confidence intervals of the mean. Schematic promoters indicate whether the synthetic or natural GSY1 promoter were used in each case.
Extended Data Figure 5 Analysis of mean gene expression dependence on time interval (continuous relative timing) between Msn2 and Mig1 pulses.
a, b, Mean expression from both synthetic (a) and natural (b) target promoters depends on the time interval between Msn2 and Mig1 pulses (that is, interval between the peak of an Msn2 pulse and the edge of the nearest Mig1 pulse). For each time interval, mean expression values were determined by integrating the area under the baseline-subtracted averaged PP7 traces, and averaging within bins of similar pulse interval. c, Specifically, Msn2 pulses were categorized on the basis of the pulse interval between Msn2 and Mig1 and the corresponding PP7 signals were averaged and their areas under curve were plotted (Methods). The pulse interval ranges from −9 to 9 min, which represents the bin centre of each 2-min bin (for example, 1 min represents the range 2 min ≥ interval > 0 min), with the 0 min interval representing overlapping events. Both >10 or <−10 min intervals represent events where Msn2 pulses were not surrounded by any Mig1 pulses within 21 min. d, e, Msn2 and Mig1 regulation are both necessary for continuous relative timing-dependent gene expression under constant glucose condition. Analysis similar to a, b was performed on synthetic Msn2- and Mig1-specific promoters (d) and natural GSY1 promoter with Msn2 or Mig1 zinc finger deletion mutants (e). Shading and error bars indicate 95% confidence intervals of the mean.
Extended Data Figure 6 Example single-cell nuclear localization traces for different constant glucose conditions.
Two single-cell traces are shown for each indicated glucose level (boxed percentage values). Cells were switched to indicated glucose level from 0.2% glucose at 110 min before time zero (that is, beginning of movie acquisition).
Extended Data Figure 7 Characterization of Msn2 and Mig1 pulses and average cross-correlation functions between Msn2 and Mig1 in individual cells across different constant glucose concentrations.
a, Pulse frequency, amplitude, and duration analysis. Single-cell traces at each glucose level were analysed and the mean frequency, amplitude and duration for both Msn2 and Mig1 were plotted. b, c, Distributions of total number of pulses per trace across glucose concentrations (b), along with corresponding fits to Poisson distributions (shown as cumulative distributions, c). Kolmogorov–Smirnov (KS) tests showed that these distributions differ significantly from Poisson distributions (P < 10−16). d, e, Analogous plots for the distributions of inter-pulse time intervals (d), and corresponding fits to exponential distributions (e). These distributions differ significantly from exponential distributions according to KS tests (P < 10−57). f, Distributions of pulse duration for Msn2 and Mig1 across glucose concentrations. g, Cross-correlation function (solid blue) of Msn2 and Mig1 nuclear localization traces, that is, cross-corr(Msn2, Mig1) (Methods). Dashed blue lines represent negative (independent) controls, calculated by scrambling the Msn2-Mig1 trace pairs within a population of cells (that is, cross-correlating Msn2 from one cell with Mig1 from another, randomly chosen, cell). Shading and error bars indicate 95% confidence intervals of the mean. The number of cells analysed in each glucose concentration: 1,511 (0.4%), 3,475 (0.2%), 2,605 (0.15%), 2,075 (0.1%), 3,034 (0.075%), 2,768 (0.05%), 1,392 (0.025%), 2,055 (0.02%), and 1,906 (0.0125%). h, Two different localization metrics show similar Msn2 and Mig1 state distributions. Top left, histogram of the intensity score for Msn2 and Mig1 shows long-tailed distributions for both proteins with peaks around zero (basal state). Insert, zoomed-in view of the tails. Top right, analogous plots for the signal proximity score also show long-tailed distributions with clear basal states. Signal proximity is the inverse of the distance-based localization metric described in the Methods section. High signal proximity indicates that the top 10 brightest pixels in the cell are close to each other. (Bottom) Signal intensity positively correlates with signal proximity for both Msn2 and Mig1, suggesting that these two independent scores show related features. This data are for cells at 0.05% glucose. Similar behaviours are observed across other glucose concentrations.
Extended Data Figure 8 Further characterization of relative pulse timing modulation under steady-state conditions.
a, Left, experimentally measured overlapping fraction (solid black) can be compared to minimum and maximum possible overlapping fractions (bottom and top dashed lines, respectively). The expected overlapping fraction for independent Msn2 and Mig1 dynamics is determined two ways: either computed from the Mig1 duty cycle (dashed black), or measured from scrambled populations (dashed red). Minimum and maximum possible fractions were calculated with the measured duty cycles of Msn2 and Mig1 pulses. Right, the ratios of measured overlapping fraction to expected overlapping fraction across glucose concentrations. b, Relative pulse timing modulation explains gene-expression dependence on glucose level for combinatorial target promoters. Black circles represent mean expression of 5 genes measured by qPCR (see Methods for normalization). Data were fit with three models, as indicated. See Methods and Supplementary Discussion for more details on binary and continuous timing models. R2 values for fits are indicated in corresponding colours. Error bars indicate s.e.m. calculated from 3 biological replicates. c, Expression data for the 5 individual genes fit to the binary timing (dashed lines; R2 values in dashed box) as well as continuous timing (solid lines; R2 values in solid box) models. d, Analysis of RNA-seq expression data across 9 glucose concentrations. The averaged expression levels from 28 of the 31 identified combinatorial targets (Extended Data Fig. 2f–k) were fit with the binary or continuous timing modulation models (left and right plots, respectively). Three genes were excluded because they did not display a monotonic dependence on glucose (YER067C-A, YKR098C, YLR109W). In this analysis, parameter b was independently estimated from an msn2 mutant at 0.2% glucose (samples collected on the same day). e, Glucose level modulates the fraction of delayed pulse timing events (see also Extended Data Fig. 4 and Supplementary Discussion). Total fractions of delayed overlapping (see Extended Data Fig. 4e, left) and delayed non-overlapping pulse events (see Extended Data Fig. 4f, left) were plotted across glucose concentrations. Expected fractions were computed from ‘scrambled’ populations where Msn2 and Mig1 dynamics are, by construction, independent. f, g, Glucose concentration also modulates relative pulse timing in a control strain without deletions of msn4 and mig2. f, Pulse characteristics of both Msn2 and Mig1 for varying glucose concentrations. g, Measured versus expected overlapping fractions across different glucose concentrations (see a) for the wild-type background that was not deleted for Msn4 and Mig2. Error bars indicate 95% confidence intervals of the mean (except for b–d). The number of cells analysed for f, g: 618 (0.4%), 541 (0.2%), 714 (0.025%), and 775 (0.0125%).
Extended Data Figure 9 Additional effects of stress level and type on transient and steady-state responses.
a, Stress level does not modulate relative pulse timing during transient responses. Averaged nuclear localization traces of Msn2–mKO2 and Mig1–mCherry during transient response to 50 mM NaCl (left) or 1.25% ethanol (right) are shown (see Fig. 2b, c). b, Additional stresses modulate relative timing during steady-state responses. Changes in pulse characteristics of both Msn2 and Mig1 in response to the addition of 100 mM NaCl or 2.5% ethanol during steady-state growth at 0.05% glucose. c, Measured (black) versus expected (grey) overlapping fractions for the same 3 conditions as in b. d, Averaged cross-correlation between Msn2 and Mig1 time traces for the same three conditions. See Supplementary Discussion for additional discussion. Shading and error bars indicate 95% confidence intervals of the mean. The number of cells analysed for b, d: 2,768 (0.05% glucose), 2,178 (0.05% glucose with 100 mM NaCl) and 2,115 (0.05% glucose with 2.5% ethanol).
Extended Data Figure 10 A role for Glc7 in active relative pulse timing modulation under constant glucose conditions and functional aspect of relative pulse timing modulation.
a, Schematic of potential mechanisms for Glc7-dependent relative pulse timing modulation (top) and construct design (bottom). Overlapping pulsing of Msn2 and Mig1 could be induced by either a common kinase/phosphatase (such as Glc7) that directly or indirectly activates both Msn2 and Mig1 localization, or by an upstream input (yellow circle) that simultaneously regulates kinases/phosphatases responsible for Msn2 and Mig1 localization. To analyse the role of GLC7 in relative pulse timing, we constructed a strain in which the normal GLC7 promoter is replaced by a copper-inducible promoter, as shown. b, qPCR characterization of the inducible GLC7 strain across three glucose concentrations. Basal copper level in the media reduced GLC7 expression to less than 50% of its wild-type level. Addition of 10 μM CuSO4 restored the expression to 110% to 140% of wild-type level. c, Changes in pulse characteristics in response to GLC7 reduction (red) and restoration (blue), compared to wild-type (black). d, Corresponding changes in pulse interval distribution. Pulse interval was calculated as the distance between the peak of a given Msn2 pulse and the peak of its closest Mig1 pulse within a 21 min window. e, Averaged nuclear localization traces of Msn2–mKO2 (green) and Mig1–mCherry (red) in response to 2.5% ethanol addition (dashed line) for the GLC7 reduction mutant. See Supplementary Discussion for additional discussion. Error bars in b indicate s.e.m. from 3 biological replicates. For c–e, shading and error bars indicate 95% confidence intervals of the mean. The number of cells analysed in the mutant strain: 671 (0.2% glucose without Cu2+), 540 (0.1% glucose without Cu2+), 719 (0.025% glucose without Cu2+), 756 (0.2% glucose with Cu2+), 643 (0.1% glucose with Cu2+), and 656 (0.025% glucose with Cu2+). f–h, Functional aspect of relative pulse timing modulation (see Supplementary Note). f, Concentration-based versus time-based regulation. Input modulates the regulator concentration (left) versus the fraction of regulator ON time (right). g, Modulation of relative pulse timing in time-based regulation results in changes in the effective protein–protein cooperativity. Increasing protein–protein cooperativity in concentration-based regulation changes the probability of co-binding of TFA and TFB (left). Increasing overlapping pulsing in time-based regulation leads to qualitatively similar changes in the probability of co-binding (right). Protein cooperativity parameter ωAB was increased from 1 to 2 for the left plots. Overlap fraction was increased from θAθB to 2 × θAθB for the right plots (ωAB = 1). KA = KB = 5 for both left and right. h, Schematic, relative pulse timing modulation affects the relative probability of simultaneous binding of two transcription factors to a target promoter (right). This effect is analogous to that generated by cooperative protein–protein interactions (left)44. Stronger protein–protein interactions or a higher overlap fraction can both increase the probability with which two transcription factors will be simultaneously bound at neighbouring sites (schematic pie charts).
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Note, Supplementary References and Supplementary Table 1. (PDF 331 kb)
Single-cell videos of Msn2-mKO2 and Mig1-mCherry in response to sudden reduction in glucose concentration
The videos and the corresponding nuclear localization traces for two cells (white circles) are shown. The switch in glucose (from 0.2% to 0.1%) is indicated by the arrow (at time = 30 min). Green and red represent Msn2-mKO2 and Mig1-mCherry, respectively. (MOV 7894 kb)
Three-color single-cell videos of the synthetic combinatorial promoter strain in response to NaCl
The videos and the corresponding nuclear localization and transcriptional activity traces for two cells (white circles) are shown. The addition of 100mM NaCl is indicated by arrow. Green, red, and blue represent Msn2-mKO2, Mig1-mCherry, and PP7-2xGFP, respectively. (MOV 2106 kb)
Three-color single-cell videos of the synthetic combinatorial promoter strain in response to ethanol
The videos and the corresponding nuclear localization and transcriptional activity traces for two cells (white circles) are shown. The addition of 2.5% ethanol is indicated by the arrow. Green, red, and blue represent Msn2-mKO2, Mig1-mCherry, and PP7-2xGFP, respectively. (MOV 2072 kb)
Three-color single-cell videos of synthetic combinatorial promoter strain under constant glucose conditions
The videos and the corresponding nuclear localization and synthetic combinatorial promoter transcriptional activity traces for two cells (white circles) are shown, at 0.05% glucose. Green, red, and blue represent Msn2-mKO2, Mig1-mCherry, and PP7-2xGFP, respectively. (MOV 11875 kb)
Three-color single-cell videos of natural GSY1 combinatorial promoter strain under constant glucose conditions
The videos and the corresponding nuclear localization and GSY1 transcriptional activity traces for two cells (white circles) are shown, at 0.05% glucose. Green, red, and blue represent Msn2-mKO2, Mig1-mCherry, and PP7-2xGFP, respectively. (MOV 11042 kb)
Rights and permissions
About this article
Cite this article
Lin, Y., Sohn, C., Dalal, C. et al. Combinatorial gene regulation by modulation of relative pulse timing. Nature 527, 54–58 (2015). https://doi.org/10.1038/nature15710
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15710
This article is cited by
-
Yeast cell fate control by temporal redundancy modulation of transcription factor paralogs
Nature Communications (2021)
-
Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line
Nature Communications (2020)
-
Identifying a stochastic clock network with light entrainment for single cells of Neurospora crassa
Scientific Reports (2020)
-
Mig1 localization exhibits biphasic behavior which is controlled by both metabolic and regulatory roles of the sugar kinases
Molecular Genetics and Genomics (2020)
-
Single-cell study links metabolism with nutrient signaling and reveals sources of variability
BMC Systems Biology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.