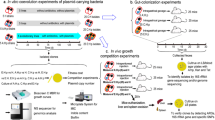
Abstract
Enterococcus faecalis is both a common commensal of the human gastrointestinal tract and a leading cause of hospital-acquired infections1. Systemic infections with multidrug-resistant enterococci occur subsequent to gastrointestinal colonization2. Preventing colonization by multidrug-resistant E. faecalis could therefore be a valuable approach towards limiting infection. However, little is known about the mechanisms E. faecalis uses to colonize and compete for stable gastrointestinal niches. Pheromone-responsive conjugative plasmids encoding bacteriocins are common among enterococcal strains3 and could modulate niche competition among enterococci or between enterococci and the intestinal microbiota. We developed a model of colonization of the mouse gut with E. faecalis, without disrupting the microbiota, to evaluate the role of the conjugative plasmid pPD1 expressing bacteriocin 21 (ref. 4) in enterococcal colonization. Here we show that E. faecalis harbouring pPD1 replaces indigenous enterococci and outcompetes E. faecalis lacking pPD1. Furthermore, in the intestine, pPD1 is transferred to other E. faecalis strains by conjugation, enhancing their survival. Colonization with an E. faecalis strain carrying a conjugation-defective pPD1 mutant subsequently resulted in clearance of vancomycin-resistant enterococci, without plasmid transfer. Therefore, bacteriocin expression by commensal bacteria can influence niche competition in the gastrointestinal tract, and bacteriocins, delivered by commensals that occupy a precise intestinal bacterial niche, may be an effective therapeutic approach to specifically eliminate intestinal colonization by multidrug-resistant bacteria, without profound disruption of the indigenous microbiota.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
GenBank/EMBL/DDBJ
Sequence Read Archive
Data deposits
The complete sequence of pPD1 was deposited in GenBank under accession number KT290268. 16S rDNA sequences generated for microbiome analyses are deposited in the NCBI-SRA archive under study accession number SRP061808 and BioProject accession number PRJNA290480.
References
Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21, 510–515 (2000)
Broaders, E., Gahan, C. G. & Marchesi, J. R. Mobile genetic elements of the human gastrointestinal tract: potential for spread of antibiotic resistance genes. Gut Microbes 4, 271–280 (2013)
Nes, I. F., Diep, D. B. & Ike, Y. in Enterococci: From Commensals to Leading Causes of Drug-Resistant Infection (eds Gilmore, M. S. et al.) Ch. 13 (Massachusetts Eye and Ear Infirmary, 2014)
Fujimoto, S., Tomita, H., Wakamatsu, E., Tanimoto, K. & Ike, Y. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J. Bacteriol. 177, 5574–5581 (1995)
Hooper, L. V., Midtvedt, T. & Gordon, J. I. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307 (2002)
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007)
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nature Rev. Immunol. 9, 313–323 (2009)
Hidron, A. I. et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29, 996–1011 (2008)
Shepard, B. D. & Gilmore, M. S. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 4, 215–224 (2002)
Paulsen, I. T. et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074 (2003)
Brandl, K. et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807 (2008)
Ubeda, C. et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010)
Dobson, A., Cotter, P. D., Ross, R. P. & Hill, C. Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78, 1–6 (2012)
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nature Rev. Microbiol. 8, 15–25 (2010)
Corr, S. C. et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl Acad. Sci. USA 104, 7617–7621 (2007)
Perez, R. H., Zendo, T. & Sonomoto, K. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact. 13 (Suppl 1), S3 (2014)
Grande Burgos, M. J., Pulido, R. P., Del Carmen Lopez Aguayo, M., Galvez, A. & Lucas, R. The cyclic antibacterial peptide enterocin AS-48: isolation, mode of action, and possible food applications. Int. J. Mol. Sci. 15, 22706–22727 (2014)
Cruz, V. L., Ramos, J., Melo, M. N. & Martinez-Salazar, J. Bacteriocin AS-48 binding to model membranes and pore formation as revealed by coarse-grained simulations. Biochim. Biophys. Acta 1828, 2524–2531 (2013)
Tomita, H., Fujimoto, S., Tanimoto, K. & Ike, Y. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179, 7843–7855 (1997)
Gálvez, A., Maqueda, M., Martinez-Bueno, M. & Valdivia, E. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against Gram-positive and Gram-negative bacteria and other organisms. Res. Microbiol. 140, 57–68 (1989)
Vesić, D. & Kristich, C. J. A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J. Bacteriol. 195, 1815–1824 (2013)
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6, 343–345 (2009)
O’Sullivan, D. J. & Klaenhammer, T. R. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59, 2730–2733 (1993)
Croswell, A., Amir, E., Teggatz, P., Barman, M. & Salzman, N. H. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect. Immun. 77, 2741–2753 (2009)
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012)
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods 10, 996–998 (2013)
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336 (2010)
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013)
Lozupone, C., Hamady, M. & Knight, R. UniFrac – an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7, 371 (2006)
Chao, A., Chazdon, R. L., Colwell, R. K. & Shen, T. J. Abundance-based similarity indices and their estimation when there are unseen species in samples. Biometrics 62, 361–371 (2006)
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948)
Oksanen, J. et al. Vegan: Community Ecology Package. R package version 2.0-10. https://cran.r-project.org/web/packages/vegan/index.html (2013)
Goslee, S. C. & Urban, D. L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19 (2007)
R. Development Core Team. R: a language and environment for statistical computing. http://www.R-project.org/ (R Foundation for Statistical Computing, 2014)
Acknowledgements
We thank I. Banla for constructing the IB1 (V583r) strain. We are grateful to J. Barbieri and the members of the Salzman and Kristich laboratories for critical review of the manuscript. We thank M.S. Gilmore for providing enterococcal strains. This work was supported by grants from the National Institutes of Health: AI057757 (N.H.S), AI097619 (N.H.S), GM099526 (N.H.S.), AI081692 (C.J.K.) and OD006447 (C.J.K).
Author information
Authors and Affiliations
Contributions
S.K., C.J.K. and N.H.S. designed and conceived the study. S.K. performed most of the experiments and the analysis; D.J.B. and M.H. assisted in the development of the colonization model. V.L. performed bioinformatics analysis. R.C. and P.B. contributed to the sequential colonization experiment. P.S. and Y.C. performed statistical analysis; S.K., C.J.K. and N.H.S. interpreted the data and wrote the manuscript. C.J.K. and N.H.S. secured funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 EF colonization.
a, C57Bl/6 mice (n = 5) were given EFr in drinking water for 14 days. Faecal samples were taken from each animal at the transition to sterile drinking water (week 0) and then weekly. EFr abundance was determined by enumeration on brain-heart infusion (BHI) agar with rifampicin. b, V583, OG1RF and JL214 strains of E. faecalis that are rifampicin resistant were fed to groups of mice (n = 5 per group). E. faecalis strains were enumerated weekly as described above. In both a and b, horizontal lines indicate geometric means and each symbol represents an individual animal. Data are representative of more than three experiments in a; in b, data are representative of more than three experiments for V583r and OG1RF and the result of one experiment for JL214.
Extended Data Figure 2 Map of plasmid pPD1.
The 59 ORFs identified in the nucleotide sequence of pPD1 are located on a circular map. Arrows indicating the direction of transcription show the ORFs. Different colours indicate coding regions for conjugation (black), bac operon (red) and maintenance or repair (blue). Hypothetical coding regions are shown in magenta. A circular plasmid map was generated using the SnapGene software (GSL Biotech; available at http://www.snapgene.com). Schematic diagrams of multiple alignments of plasmids were produced by manually realigning the linear plasmid maps generated by the SnapGene Viewer.
Extended Data Figure 3 pPD1 enhances E. faecalis competition for an intestinal niche.
a, Bacteriocin assay by the soft agar method, with EFr + pPD1::ΔbacAB bacA-E+, EFr + pPD1 and EFr + pPD1::ΔbacAB spotted on a lawn of susceptible E. faecalis. Mice (n = 5 per group) were given EFr, EFr + pPD1, EFr + pPD1::ΔbacAB or sterile drinking water for 14 days, after which all mice were given sterile water. b, c, One week (b) and four weeks (c) after withdrawal of E. faecalis from the drinking water, faecal samples were collected and the abundance of total enterococci was determined by enumeration on m-Enterococcus selective agar (Ent agar). Laboratory strains of E. faecalis were enumerated using Ent agar with rifampicin (Rf) or BHI agar with rifampicin. d, e, At the end of week 4, animals were euthanized and abundance of E. faecalis was determined in the distal small intestine (d) and large intestine (e) (also see Fig. 1e). The results shown are representative of two biologically independent experiments. In b–e, horizontal lines indicate geometric means and each symbol represents an individual animal.
Extended Data Figure 4 EF + pPD1 but not EFr + pCF10 dominates the intestinal enterococcal population.
pCF10 is a well-studied pheromone-inducible conjugative plasmid of E. faecalis that encodes resistance to tetracycline but does not encode a known bacteriocin determinant. a, EFr (n = 3 mice, no plasmid), EFr + pPD1 (n = 3 mice, pPD1) or EFr + pCF10, (n = 5 mice, pCF10) was added to the drinking water for 14 days and then replaced by sterile drinking water. Faecal samples were taken from each animal at the transition to sterile drinking water (week 0) and then weekly. E. faecalis abundance was determined by enumeration on BHI agar with rifampicin. An exponential decay model is used to fit the data and there are significant differences between the groups pCF10 (red) and pPD1 (blue) (P = 0.012), as well as between the pPD1-containing and plasmid-free groups (black) (P = 0.007). b, Four weeks after withdrawal of E. faecalis from the drinking water, animals were euthanized and abundance of EFr + pCF10 was determined in the distal small intestine (DSI), caecum and large intestine (LI). Mice colonized with EFr + pCF10 maintained long-term faecal shedding of EFr + pCF10 similar to EFr and persistent colonization throughout the gastrointestinal tract. c, d, Abundance of enterococci in the faeces was determined by enumeration on m-Enterococcus agar (Ent agar), Ent agar with rifampicin or BHI agar with rifampicin at week 1 (c) and week 4 (d). Unlike EFr + pPD1, which dominated the enterococcal niche in the gastrointestinal tract (Fig. 1e), EFr + pCF10 did not outcompete the indigenous enterococci, colonizing at levels comparable to EFr. In b–d, horizontal lines indicate geometric means and each symbol represents an individual animal; data were obtained from one experiment.
Extended Data Figure 5 Microbiome analysis.
a, NMDS ordination of control (no EF treatment) and E. faecalis samples as separated by the Bray–Curtis beta diversity metric. Control (n = 5 mice) and E. faecalis samples (n = 5 mice) are intermixed. No significant difference in beta diversity was seen between the two groups. Adonis P-value = 0.298. Samples are connected with lines to help visualize grouping. b, Analysis between E. faecalis and EF + pPD1 samples at the level of operational taxonomic unit suggests changes in the abundance of four bacterial genera, in particular for Deferribacteraceae; Mucispirillum and Lachnospiraceae; Incertae sedis (P = 0.0001 and P = 0.005, respectively; heteroscedastic two-sided Student’s t-test). Bacteria belonging to these two genera were tenfold and twofold lower, respectively, in EF + pPD1-colonized mice. (Note that the magnitude of change is shown in log10 scale.) Changes in Defluviitaleaceae; Incertae sedis and Lachnospiraceae; Blautia were not as pronounced (P = 0.03; heteroscedastic two-sided Student’s t-test). However, analyses at the family taxonomy level suggest that the change in Deferribacteraceae was statistically significant.
Extended Data Figure 6 Reciprocal experiment for data from Fig. 2a, d and g.
Groups of mice (n = 5 mice per group) were given mixtures of EFr + pPD1 and EFs in drinking water at ratios of 10%/90% (a), 50%/50% (b) and 90%/10% (c), respectively. Faecal samples were obtained at the transition to sterile drinking water (week 0) and then weekly. Abundance of each E. faecalis strain in the faeces was determined by enumeration on BHI agar with rifampicin and BHI agar with spectinomycin. Abundance of EFr + pPD1 is indicated by open squares; abundance of EFs is indicated by open circles. Each symbol represents an individual animal. The differences between the two groups at each week were compared using a non-parametric Wilcoxon test. In a, no P-values were significant, in b, P < 0.005 and in c, P = 0.0122 (week 0) and P = 0.0075 (week 1–4). An exponential decay model is used for fitting the data in a and b. The results in a–c are from one experiment.
Extended Data Figure 7 pPD1-associated competition in vitro.
Three independent cultures were carried out with various mixed populations of EFr (–) and EFs + pPD1 (+) or EFr (–) and EFs + pPD1::ΔbacAB (+ΔbacAB) in 10 ml BHI broth at ratios of 90%/10% (a, d), 50%/50% (b, e) and 10%/90% (c, f). Samples for serial dilution were taken at 0, 2, 4, 6 and 24 h after the start of the experiment. Abundance of each E. faecalis strain in faeces was determined by enumeration on BHI agar with rifampicin and BHI agar with spectinomycin. Evidence of conjugation was observed in in vitro co-cultures of a and b only by screening for transconjugants (EFr + pPD1) via colony PCR. Open squares represent abundance of EFs + pPD1 (a–c) or EFs + pPD1::ΔbacAB (d–f). Open circles represent abundance of EFr in all panels. Data are representative of two biologically independent experiments.
Extended Data Figure 8 Complementation of bac-21 production restores colonization phenotype by providing a competitive advantage.
Bacteriocin activity was restored upon ectopic expression of bacA to bacE (from pAM401) in EF + pPD1::ΔbacAB but not in E. faecalis lacking pPD1, indicating that the distal part of the bac operon (bacF to bacI) is necessary for bacteriocin expression and that the bacAB in-frame deletion is not polar on downstream genes. Mice (n = 5) were given EFr + pPD1::ΔbacAB bacABCDE+ as described in the methods and abundance was determined by enumeration on m-Enterococcus (Ent) agar, m-Enterococcus agar plus rifampicin (Ent-Rif) or BHI agar with rifampicin (BHI-Rif). The presence of pAM401A::bacABCDE + (complementing plasmid) was determined by enumerating CFU on BHI agar with rifampicin and chloramphenicol (BHI-Rif Cm). Faecal samples were obtained at week 1 (a) and week 4 (b) after transition to sterile drinking water. Horizontal lines indicate geometric means. Each symbol represents an individual animal and data are representative of two biologically independent experiments. EFs + pPD1::ΔbacAB bacABCDE + stably colonized the gastrointestinal tract (a); however, in the absence of chloramphenicol selection, pAM401::bacABCDE was gradually lost from the population (b). Over time, loss of pAM401::bacABCDE resulted in the complemented strain reverting to the bacteriocin-defective ΔbacAB strain, with the loss of bacteriocin activity. Nevertheless, this strain persisted in the gut, suggesting that bac-21 was essential for clearing a niche for E. faecalis; once cleared, E. faecalis uses other mechanisms to maintain colonization.
Extended Data Figure 9 EFr levels are not altered by sequential colonization with EFs + pPD1.
Groups of mice (n = 5 mice per group) were given EFr in drinking water for two weeks and subsequently challenged with EFs + pPD1 in drinking water for another two weeks (starting from week –2) before transition to sterile water (week 0). Faecal samples were obtained weekly to enumerate the abundance of EFr (a) and EFs + pPD1 (b) on BHI agar with rifampicin and spectinomycin, respectively. Each symbol represents an individual animal and data are from one experiment.
Extended Data Figure 10 Conjugation frequency of pPD1 between the laboratory strain (EF + pPD1) and indigenous enterococci in vitro.
To understand conjugation dynamics between non-isogeneic species of enterococci, we investigated indigenous enterococcal transconjugants in mice that were colonized with EFr + pPD1 (Fig. 1e and Extended Data Fig. 3b–e). However, we were unable to detect rifampicin-sensitive enterococci from the faecal sample at week 4 (Extended Data Fig. 3c). At week 1, only nine clones of rifampicin-sensitive enterococci (out of 730 enterococci) were isolated from three mice in a group of five (Extended Data Fig. 3b). Bacteriocin assays and probing for the bacA gene sequence confirmed that six of the nine clones were transconjugants and were bac-21 positive. 16S rDNA gene sequencing of these nine clones showed notable similarities to E. faecalis 16S rDNA. To understand the reason for the low conjugation frequency between indigenous enterococci and the laboratory strain of EFr + pPD1, in vitro conjugation experiments were performed to assess the frequency of plasmid transfer. Ten new clones of indigenous enterococci were isolated from the faeces of five mice (two clones per mouse that were not colonized with any laboratory strain) by culturing on m-Enterococcus agar. EFr + pPD1 was mixed with each of the ten indigenous enterococci clones in BHI broth at a ratio of 1:9. Samples for serial dilution were taken at the start of the culture (a) and 24 h after the start of the experiment (b). Abundance of total enterococci was determined using m-Enterococcus agar and BHI agar with rifampicin for EFr + pPD1. Data are representative of three biologically independent experiments. In vitro bacteriocin assays revealed that EFr + pPD1 is capable of killing most of the non-pPD1-containing indigenous enterococci strains (not shown). In vitro conjugation assays between individual indigenous enterococci clones and EFr + pPD1 led to three observations: first, 4 out of 10 clones were susceptible to bac-21 and were eliminated by EFr + pPD1 (no. 3, 4, 8 and 9); second, 6 out of 10 were immune to bac-21 killing (no. 1, 2, 5, 6, 7 and 10); third, probing for bacA provided evidence for pPD1-containing indigenous enterococci transconjugants in 4 out of the 6 immune indigenous enterococci clones. The two clones that failed to conjugate and were resistant to bac-21 killing in the mixed culture experiment were also resistant to EFr + pPD1 on bacteriocin assay plates. The mechanism for resistance of these two clones is not clear; however, they might have harboured cross-resistance traits. Error bars represent s.e.m.
Supplementary information
Supplementary Table 1
This file contains comparisons between (a) amino acid sequences, deduced from the nucleotide sequences of putative coding regions and (b) nucleotide sequence identities between AS-48 gene cluster of E. faecalis S-48 (GenBank accession number Y12234 and AJ438950) and Bac-21 gene cluster of E. faecalis CK135 pPD1 (GenBank accession number KT290268). (XLSX 46 kb)
Supplementary Table 2
This file contains a list of open reading frames (ORFs) identified in the pPD1 sequence. (XLSX 43 kb)
Supplementary Table 3
This file lists the frequency of transconjugants that harbor pPD1 in the GI tract. Numerator indicates the number of transconjugants that harbor pPD1, and denominator indicates the total number of clones screened through colony PCR in each mouse (m). (XLSX 42 kb)
Supplementary Table 4
This file contains the list of susceptible strains of E. faecalis and E. faecium towards Bac-21 produced by EF. (XLSX 39 kb)
Supplementary Table 5
This file contains the list of strains and plasmids used in this study. (XLSX 38 kb)
Supplementary Table 6
This file contains sequences of primers used in this study. (XLSX 44 kb)
Supplementary Notes
This file contains the Supplementary References for Supplementary Table 4 (see references 37 and 38) and Supplementary Table 5 (see references 39–45). (PDF 108 kb)
Rights and permissions
About this article
Cite this article
Kommineni, S., Bretl, D., Lam, V. et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722 (2015). https://doi.org/10.1038/nature15524
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15524
This article is cited by
-
Increased Intestinal Permeability: An Avenue for the Development of Autoimmune Disease?
Exposure and Health (2024)
-
Circular and L50-like leaderless enterocins share a common ABC-transporter immunity gene
BMC Genomics (2023)
-
A pilot study demonstrating the impact of surgical bowel preparation on intestinal microbiota composition following colon and rectal surgery
Scientific Reports (2022)
-
Microbiome-mediated fructose depletion restricts murine gut colonization by vancomycin-resistant Enterococcus
Nature Communications (2022)
-
Genome editing of probiotic bacteria: present status and future prospects
Biologia (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.