Abstract
Acetyl-CoA carboxylase (ACC) has crucial roles in fatty acid metabolism and is an attractive target for drug discovery against diabetes, cancer and other diseases1,2,3,4,5,6. Saccharomyces cerevisiae ACC (ScACC) is crucial for the production of very-long-chain fatty acids and the maintenance of the nuclear envelope7,8. ACC contains biotin carboxylase (BC) and carboxyltransferase (CT) activities, and its biotin is linked covalently to the biotin carboxyl carrier protein (BCCP). Most eukaryotic ACCs are 250-kilodalton (kDa), multi-domain enzymes and function as homodimers and higher oligomers. They contain a unique, 80-kDa central region that shares no homology with other proteins. Although the structures of the BC, CT and BCCP domains and other biotin-dependent carboxylase holoenzymes are known1,9,10,11,12,13,14, there is currently no structural information on the ACC holoenzyme. Here we report the crystal structure of the full-length, 500-kDa holoenzyme dimer of ScACC. The structure is remarkably different from that of the other biotin-dependent carboxylases. The central region contains five domains and is important for positioning the BC and CT domains for catalysis. The structure unexpectedly reveals a dimer of the BC domain and extensive conformational differences compared to the structure of the BC domain alone, which is a monomer. These structural changes reveal why the BC domain alone is catalytically inactive and define the molecular mechanism for the inhibition of eukaryotic ACC by the natural product soraphen A15,16 and by phosphorylation of a Ser residue just before the BC domain core in mammalian ACC. The BC and CT active sites are separated by 80 Å, and the entire BCCP domain must translocate during catalysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Tong, L. Structure and function of biotin-dependent carboxylases. Cell. Mol. Life Sci. 70, 863–891 (2013)
Waldrop, G. L., Holden, H. M. & St Maurice, M. The enzymes of biotin dependent CO2 metabolism: what structures reveal about their reaction mechanisms. Protein Sci. 21, 1597–1619 (2012)
Cronan, J. E., Jr & Waldrop, G. L. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41, 407–435 (2002)
Polyak, S. W., Abell, A. D., Wilce, M. C. J., Zhang, L. & Booker, G. W. Structure, function and selective inhibition of bacterial acetyl-CoA carboxylase. Appl. Microbiol. Biotechnol. 93, 983–992 (2012)
Abramson, H. N. The lipogenesis pathway as a cancer target. J. Med. Chem. 54, 5615–5638 (2011)
Wakil, S. J. & Abu-Elheiga, L. A. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 50 (Suppl), S138–S143 (2009)
Schneiter, R. et al. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16, 7161–7172 (1996)
Hoja, U., Wellein, C., Greiner, E. & Schweizer, E. Pleiotropic phenotype of acetyl-CoA-carboxylase-defective yeast cells. Eur. J. Biochem. 254, 520–526 (1998)
St Maurice, M. et al. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science 317, 1076–1079 (2007)
Xiang, S. & Tong, L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nature Struct. Mol. Biol. 15, 295–302 (2008)
Huang, C. S. et al. Crystal structure of the α6β6 holoenzyme of propionyl-coenzyme A carboxylase. Nature 466, 1001–1005 (2010)
Huang, C. S., Ge, P., Zhou, Z. H. & Tong, L. An unanticipated architecture of the 750-kDa α6β6 holoenzyme of 3-methylcrotonyl-CoA carboxylase. Nature 481, 219–223 (2012)
Fan, C., Chou, C.-Y., Tong, L. & Xiang, S. Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction. J. Biol. Chem. 287, 9389–9398 (2012)
Tran, T. H. et al. Structure and function of a single-chain, multi-domain long-chain acyl-CoA carboxylase. Nature 518, 120–124 (2015)
Weatherly, S. C., Volrath, S. L. & Elich, T. D. Expression and characterization of recombinant fungal acetyl-CoA carboxylase and isolation of a soraphen-binding domain. Biochem. J. 380, 105–110 (2004)
Shen, Y., Volrath, S. L., Weatherly, S. C., Elich, T. D. & Tong, L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol. Cell 16, 881–891 (2004)
Zhang, H., Yang, Z., Shen, Y. & Tong, L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science 299, 2064–2067 (2003)
Chapman-Smith, A., Forbes, B. E., Wallace, J. C. & Cronan, J. E., Jr Covalent modification of an exposed surface turn alters the global conformation of the biotin carrier domain of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 272, 26017–26022 (1997)
Solbiati, J., Chapman-Smith, A. & Cronan, J. E., Jr Stabilization of the biotinoyl domain of Escherichia coli acetyl-CoA carboxylase by interactions between the attached biotin and the protruding “thumb” structure. J. Biol. Chem. 277, 21604–21609 (2002)
Hung, C. L. et al. Crystal structure of Helicobacter pylori formamidase AmiF reveals a cysteine-glutamate-lysine catalytic triad. J. Biol. Chem. 282, 12220–12229 (2007)
Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008)
Raymer, B. et al. Synthesis and characterization of a BODIPY-labeled derivative of Soraphen A that binds to acetyl-CoA carboxylase. Bioorg. Med. Chem. Lett. 19, 2804–2807 (2009)
Cho, Y. S. et al. Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK. Biochem. Biophys. Res. Commun. 391, 187–192 (2010)
Waldrop, G. L., Rayment, I. & Holden, H. M. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry 33, 10249–10256 (1994)
Shen, Y., Chou, C.-Y., Chang, G.-G. & Tong, L. Is dimerization required for the catalytic activity of bacterial biotin carboxylase? Mol. Cell 22, 807–818 (2006)
Chou, C.-Y., Yu, L. P. C. & Tong, L. Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism. J. Biol. Chem. 284, 11690–11697 (2009)
Harwood, H. J., Jr et al. Isozyme-nonselective N-substituted bipiperidylcarboxamide acetyl-CoA carboxylase inhibitors reduce tissue malonyl-CoA concentrations, inhibit fatty acid synthesis, and increase fatty acid oxidation in cultured cells and in experimental animals. J. Biol. Chem. 278, 37099–37111 (2003)
Zhang, H., Tweel, B., Li, J. & Tong, L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase in complex with CP-640186. Structure 12, 1683–1691 (2004)
Chou, C.-Y. & Tong, L. Structural and biochemical studies on the regulation of biotin carboxylase by substrate inhibition and dimerization. J. Biol. Chem. 286, 24417–24425 (2011)
Smith, A. C. & Cronan, J. E. Dimerization of the bacterial biotin carboxylase subunit is required for acetyl coenzyme A carboxylase activity in vivo. J. Bacteriol. 194, 72–78 (2012)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Blanchard, C. Z., Lee, Y. M., Frantom, P. A. & Waldrop, G. L. Mutations at four active site residues of biotin carboxylase abolish substrate-induced synergism by biotin. Biochemistry 38, 3393–3400 (1999)
Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999)
Acknowledgements
We thank M. Bush, C.-Y. Chou, Y. Shen, L. Yu and H. Zhang for carrying out initial studies in this project; R. Jackimowicz, B. Nolan, N. Whalen, A. Heroux and H. Robinson for access to the X29A and X25 beamlines at the NSLS; S. Banerjee, K. Perry, R. Rajashankar, J. Schuermann, N. Sukumar for access to NE-CAT 24-C and 24-E beamlines at the Advanced Photon Source; W. Rice and E. Eng at New York Structural Biology Center for assistance with electron microscopy. The in-house X-ray diffraction instrument was purchased with an NIH grant to L.T. (S10OD012018). This research was supported in part by a grant from the NIH (R01DK067238) to L.T.
Author information
Authors and Affiliations
Contributions
J.W. carried out protein expression, purification, crystallization, data collection, structure determination and refinement, site-directed mutagenesis and enzymatic assays. L.T. initiated the project, supervised the entire research, and analysed the results. J.W. and L.T. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Sequence alignment of the N-terminal region of eukaryotic, single-chain ACCs.
The region includes BC, BT and BCCP domains (indicated). The secondary structure elements in the ScACC holoenzyme structure are shown. The site of phosphorylation in HsACC1 (Ser80) is indicated with a star. This site does not exist in ScACC. ScACC, Saccharomyces cerevisiae ACC; TaACC, Triticum aestivum (wheat) ACC; DmACC, Drosophila melanogaster ACC; HsACC1, Homo sapiens ACC1. Modified from an output from ESPript41.
Extended Data Figure 2 Sequence alignment of the central region of eukaryotic, single-chain ACCs.
The AC1–AC5 domains are indicated. Predicted secondary structure elements in HsACC1 are also shown, and they generally match those in the structure of ScACC. The helices in AC1–AC3 are numbered consecutively. Three sites of phosphorylation in HsACC1 are indicated with stars.
Extended Data Figure 3 Sequence alignment of the C-terminal region of eukaryotic, single-chain ACCs.
The region includes N and C domains of CT (indicated).
Extended Data Figure 4 Electron density for various regions of the ScACC holoenzyme structure.
a, Schematic of the two-step reaction in the catalysis by biotin-dependent carboxylases. Biotin is carboxylated in the BC active site, and then translocates to the CT active site where the substrate is carboxylated. The longest distance from the N1′ atom of biotin to the Cα atom of the Lys residue covalently attached to biotin is ∼16 Å, giving a reach of ∼30 Å for biotin (swinging-arm model). The actual distance between the BC and CT active sites are larger than 30 Å, suggesting that BCCP needs to translocate in addition to the biotin (swinging-domain model). b, 2Fo − Fc electron density for the helical hairpin insert (α8–α9) of domain AC1 at 3.2 Å resolution, contoured at 1σ. c, 2Fo − Fc electron density for the β-sheet of domain AC5. d, 2Fo − Fc electron density for the central helix (αV) of the BT domain. e, Omit Fo − Fc electron density for CoA at 3.2 Å resolution, contoured at 2.5σ. f, Omit Fo − Fc electron density for biotin at 3.2 Å resolution.
Extended Data Figure 5 Overall structure of the ScACC holoenzyme.
a, A large channel in the centre of the ScACC holoenzyme dimer. The view is related to that of Fig. 1b by an ∼30° rotation around the vertical axis. b, Overlay of the structures of the two protomers of ScACC holoenzyme dimer. One protomer is shown in colour and other in grey. The overlay is based on the CT domain. Differences in the orientations of the other domains are indicated, as well as the r.m.s. distance for their equivalent Cα atoms. The arrow points to conformational differences in the insert domain of CT, linked to differences in the BCCP binding mode. c, Overlay of the BT domain of ScACC (in orange) and the BT domain of PCC (in grey). The ‘hook’, connecting the end of the helix (αV) to the first strand of the β-barrel (β22), is labelled. The last three strands of the β-barrel (β27–β29) are splayed further away from the central helix in ScACC compared to PCC. d, Interactions between the hook of the BT domain and the β4A–β4B loop from the C domain of CT, an inserted segment that projects away from the rest of the domain. e, The BCCP–AC1 linker has hydrophobic interactions with the top of one side of the BT domain β-barrel. The disordered segment of the linker is indicated with the blue line.
Extended Data Figure 6 Domains AC4 and AC5 share a common backbone fold.
a, Structure of AC4 domain of ScACC. b, Topological drawing of AC4 domain. c, Structure of AC5 domain. d, Topological drawing of AC5 domain. e, Overlay of the structures of AC5 domain (magenta) and formamidase (grey). Formamidase has a four-layered αββα structure, and AC5 matches only half of the structure.
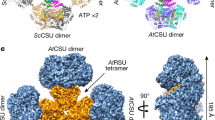
Extended Data Figure 7 Structure of the BC domain dimer of ScACC.
a, Interactions in the BC dimer interface. The BC domain of protomer 1 is in red, and that of protomer 2 in pink. b, Residues in the BC dimer interface (in red) of ScACC and E. coli BC (EcBC) are weakly conserved among the biotin-dependent carboxylases. MapLCC, long-chain acyl-CoA carboxylase of Mycobacterium avium subspecies tuberculosis. c, A BC domain dimer is constructed by superposing the structure of BC domain alone (in complex with soraphen A, green) onto that of the BC dimer. Regions of steric clashes between the two monomers are highlighted in light blue. d, Close-up view of the binding site of biotin. The β19–β20 loop in the structure of BC domain alone (grey) clashes with biotin (red arrow), and cannot interact with the amide group of the biotin linkage (red dashed lines). e, Close-up view of the soraphen A (green) binding site. The movement of strands β19 and β20 from the structure of BC domain alone in complex with soraphen A (grey) is indicated with the arrows. Trp487 side chain in the holoenzyme structure clashes with soraphen A as well as the phosphorylated peptide segment containing pSer222 (light blue).
Extended Data Figure 8 Structure of the CT domain dimer of ScACC.
a, Overlay of the CT domain dimer in the ScACC holoenzyme (cyan and yellow for N and C domains of protomer 1, light cyan and light yellow for protomer 2) in complex with CoA (black) with the CT domain alone in complex with CoA (grey). Biotin is shown in black and BCCP is omitted for clarity. The bound position of the CP-640186 inhibitor (gold) is shown for reference. A conformational change for the insert domain at the top is due to the binding of BCCP in the holoenzyme, while the insert domain at the bottom shows essentially no change because the BCCP–biotin is not bound as deeply into this active site. b, Overlay of the CT active site (cyan and yellow) of ScACC holoenzyme with that of CT alone in complex with CoA (grey). The CP-640186 inhibitor (gold) clashes with the bound position of biotin (black). The thiol group of CoA in the holoenzyme complex is 4.3 Å from the N1 atom of biotin (dashed line in blue). The thiol group of CoA in the CT domain alone complex is in a different position, likely due to the absence of biotin in the active site.
Extended Data Figure 9 Comparisons of the structures of ACC central region alone with that in the holoenzyme.
a, Overlay of AC1–AC2 in the holoenzyme (colour) with AC1–AC2 alone (yellow and grey). The first helix is swapped between two monomers in the structure of AC1–AC2 alone, and the two-fold axis of that dimer is indicated with the black line. b, Overlay of AC3–AC5 in the holoenzyme (colour) with AC3–AC5 alone (grey), based on AC3–AC4. A large difference is seen for the orientation of AC5. c, Overlay of BT–BCCP–AC1–AC5 in the holoenzyme (colour) with these domains alone (grey), based on AC1–AC2. Large differences are seen for BT, BCCP and AC3–AC5. d, Overlay of BT–BCCP–AC1–AC5 in the holoenzyme (colour) with these domains alone (grey), based on AC3–AC4. Large differences are seen for BT, BCCP and AC1–AC2, although AC5 has essentially the same position.
Supplementary information
Conformational changes in the BC domain near the dimer interface
Video morphing the structure of the BC domain alone (gray) to that in the holoenzyme (red), illustrating the large conformational changes in the BC dimer interface. The inhibitor soraphen A (green) and the side chain of Trp487 are shown as stick models. (MOV 916 kb)
Conformational changes in the BC domain near the active site
Video morphing the structure of the BC domain alone (gray) to that in the holoenzyme (red), illustrating the large conformational changes in the BC active site region. ADP (green) and biotin (black), observed bound to the active site of E. coli BC subunit, are shown as stick models. (MOV 908 kb)
Rights and permissions
About this article
Cite this article
Wei, J., Tong, L. Crystal structure of the 500-kDa yeast acetyl-CoA carboxylase holoenzyme dimer. Nature 526, 723–727 (2015). https://doi.org/10.1038/nature15375
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15375
This article is cited by
-
Algorithm-aided engineering of aliphatic halogenase WelO5* for the asymmetric late-stage functionalization of soraphens
Nature Communications (2022)
-
CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase
Nature Communications (2022)
-
Lipogenesis inhibitors: therapeutic opportunities and challenges
Nature Reviews Drug Discovery (2022)
-
WZ66, a novel acetyl-CoA carboxylase inhibitor, alleviates nonalcoholic steatohepatitis (NASH) in mice
Acta Pharmacologica Sinica (2020)
-
A novel multidomain acyl-CoA carboxylase in Saccharopolyspora erythraea provides malonyl-CoA for de novo fatty acid biosynthesis
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.