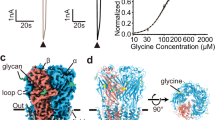
Abstract
Neurotransmitter-gated ion channels of the Cys-loop receptor family are essential mediators of fast neurotransmission throughout the nervous system and are implicated in many neurological disorders. Available X-ray structures of prokaryotic and eukaryotic Cys-loop receptors provide tremendous insights into the binding of agonists, the subsequent opening of the ion channel, and the mechanism of channel activation1,2,3,4,5,6,7,8. Yet the mechanism of inactivation by antagonists remains unknown. Here we present a 3.0 Å X-ray structure of the human glycine receptor-α3 homopentamer in complex with a high affinity, high-specificity antagonist, strychnine. Our structure allows us to explore in detail the molecular recognition of antagonists. Comparisons with previous structures reveal a mechanism for antagonist-induced inactivation of Cys-loop receptors, involving an expansion of the orthosteric binding site in the extracellular domain that is coupled to closure of the ion pore in the transmembrane domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hilf, R. J. & Dutzler, R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008)
Hilf, R. J. & Dutzler, R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 (2009)
Bocquet, N. et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009)
Sauguet, L. et al. Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl Acad. Sci. USA 111, 966–971 (2014)
Hibbs, R. E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011)
Miller, P. S. & Aricescu, A. R. Crystal structure of a human GABAA receptor. Nature 512, 270–275 (2014)
Hassaine, G. et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281 (2014)
Althoff, T., Hibbs, R. E., Banerjee, S. & Gouaux, E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 (2014)
Legendre, P. The glycinergic inhibitory synapse. Cell. Mol. Life Sci. 58, 760–793 (2001)
Lynch, J. W. Molecular structure and function of the glycine receptor chloride channel. Physiol. Rev. 84, 1051–1095 (2004)
Lynch, J. W. & Callister, R. J. Glycine receptors: a new therapeutic target in pain pathways. Curr. Opin. Investig. Drugs 7, 48–53 (2006)
Bode, A. & Lynch, J. W. The impact of human hyperekplexia mutations on glycine receptor structure and function. Mol. Brain 7, 2 (2014)
Schofield, P. R. et al. Sequence and functional expression of the GABAA receptor shows a ligand-gated receptor super-family. Nature 328, 221–227 (1987)
Langosch, D., Thomas, L. & Betz, H. Conserved quaternary structure of ligand-gated ion channels: the postsynaptic glycine receptor is a pentamer. Proc. Natl Acad. Sci. USA 85, 7394–7398 (1988)
Brejc, K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001)
Celie, P. H. et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 41, 907–914 (2004)
Hansen, S. B. et al. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 24, 3635–3646 (2005)
Billen, B. et al. Molecular actions of smoking cessation drugs at α4β2 nicotinic receptors defined in crystal structures of a homologous binding protein. Proc. Natl Acad. Sci. USA 109, 9173–9178 (2012)
Moraga-Cid, G. et al. Allosteric and hyperekplexic mutant phenotypes investigated on an α1 glycine receptor transmembrane structure. Proc. Natl Acad. Sci. USA 112, 2865–2870 (2015)
Rajendra, S., Lynch, J. W. & Schofield, P. R. The glycine receptor. Pharmacol. Ther. 73, 121–146 (1997)
Laube, B., Maksay, G., Schemm, R. & Betz, H. Modulation of glycine receptor function: a novel approach for therapeutic intervention at inhibitory synapses? Trends Pharmacol. Sci. 23, 519–527 (2002)
Grenningloh, G. et al. Alpha subunit variants of the human glycine receptor: primary structures, functional expression and chromosomal localization of the corresponding genes. EMBO J. 9, 771–776 (1990)
Jensen, A. A., Gharagozloo, P., Birdsall, N. J. & Zlotos, D. P. Pharmacological characterisation of strychnine and brucine analogues at glycine and α7 nicotinic acetylcholine receptors. Eur. J. Pharmacol. 539, 27–33 (2006)
Mohsen, A. M., Heller, E., Holzgrabe, U., Jensen, A. A. & Zlotos, D. P. Structure–activity relationships of strychnine analogs at glycine receptors. Chem. Biodivers. 11, 1256–1262 (2014)
Grudzinska, J. et al. The β subunit determines the ligand binding properties of synaptic glycine receptors. Neuron 45, 727–739 (2005)
Brams, M. et al. A structural and mutagenic blueprint for molecular recognition of strychnine and d-tubocurarine by different Cys-loop receptors. PLoS Biol. 9, e1001034 (2011)
Becker, C. M., Hoch, W. & Betz, H. Glycine receptor heterogeneity in rat spinal cord during postnatal development. EMBO J. 7, 3717–3726 (1988)
Vandenberg, R. J., French, C. R., Barry, P. H., Shine, J. & Schofield, P. R. Antagonism of ligand-gated ion channel receptors: two domains of the glycine receptor α subunit form the strychnine-binding site. Proc. Natl Acad. Sci. USA 89, 1765–1769 (1992)
Calimet, N. et al. A gating mechanism of pentameric ligand-gated ion channels. Proc. Natl Acad. Sci. USA 110, E3987–E3996 (2013)
Yu, R. et al. Agonist and antagonist binding in human glycine receptors. Biochemistry 53, 6041–6051 (2014)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007)
Schuttelkopf, A. W. & van Aalten, D. M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D 60, 1355–1363 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996)
DeLano, W. L. The PyMOL molecular graphics system (DeLano Scientific, 2002)
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007)
Jensen, A. A. & Kristiansen, U. Functional characterisation of the human α1 glycine receptor in a fluorescence-based membrane potential assay. Biochem. Pharmacol. 61, 1789–1799 (2004)
Acknowledgements
We thank G. Ranieri and R. Walter at Shamrock Structures and the staff at beamlines 08-ID at the Canadian Light Source and 22-ID at the Advanced Photon Source for data collection. We are grateful to Z. Wang and J. Gingras for reviewing the manuscript.
Author information
Authors and Affiliations
Contributions
The authors have jointly contributed to project design, data analysis and manuscript preparation. P.L.S. performed initial construct design and purification experiments, structure solution, model building, and structural analysis; X.H. performed protein purifications, crystallization, model building, and structural analysis; H.C. performed cloning and expression experiments; K.M. performed SPR and ITC binding studies; S.S. performed functional testing; P.L.S. and X.H. wrote the manuscript with help from K.M., S.S., and H.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Strychnine binding to GlyRα3cryst in detergent micelles.
a, Binding thermodynamics and stoichometry of strychnine to GlyRα3cryst analysed by ITC. The individual peaks from titrations are integrated and presented in a Wiseman plot. An appropriate binding model is chosen and the isotherm is then fitted to yield the binding enthalpy ΔH, the dissociation constant Kd, and the stoichiometry n. b, Binding kinetics of strychnine to GlyRα3cryst measured by SPR spectroscopy.
Extended Data Figure 2 Glycine dose–response to GlyRα3cryst in HEK293T cells.
Glycine dose–response curve for BacMam baculovirus-transduced HEK293T cells expressing either wild-type human GlyRα3 (GlyRα3, filled circles) or GlyRα3cryst (open circles) measured by membrane potential dye assay (see Methods for details). Each data point represents a value of n = 4–6 and normalized to a maximum glycine response observed at 2 mM glycine concentration. Glycine EC50 for GlyRα3 was calculated at 150 ± 10.6 µM (n = 4, 95% confidence interval) and GlyRα3cryst at 16.4 ± 1.2 µM (n = 5, 95% confidence interval).
Extended Data Figure 3 Architecture of the GlyRα3 bound to strychnine and crystallographic packing of GlyRα3cryst.
a, The GlyRα3–strychnine complex viewed from the extracellular side of the membrane down the pore axis, perpendicular to the membrane. Strychnine is bound at the interface between subunits. b, The GlyRα3–strychnine complex viewed from the intracellular side down the pore axis. The M2 helices are shown lining the pore. c, d, Packing of the GlyRα3–strychnine complex. Receptor in the asymmetric unit is coloured by subunit, with subunit A in pale green and subunit B in cyan. Strychnine molecules are shown as spheres and are coloured to match their associated principal subunit. Symmetry-related receptors are coloured grey. The interface between subunits A and B is completely exposed to solvent in the crystal lattice, and this interface was used in the making of all figures for this paper to avoid any potential crystal-packing artefacts.
Extended Data Figure 4 Strychnine binding to GlyRα3.
a, b, Two views of the strychnine-binding pocket showing 2Fo – Fc omit electron density maps. The principal subunit is coloured pale green, the complementary subunit is cyan, and strychnine is grey. Strychnine was omitted from map calculations. Map is contoured at 1.0σ. The hydrogen bond between the backbone carbonyl of Phe159 and the tertiary nitrogen of strychnine is shown as a dotted line. c, d, Same views and representations as in a and b, but Fo – Fc omit electron density is shown. Contour level of map is 3.0σ. e, Chemical structure of strychnine. Ring numbering is denoted by red Roman numerals. Carbon and nitrogen historic numbering conventions are denoted by blue numbers. f, Solvent-accessible surface area of the complementary side of the binding interface coloured by electrostatic potential. The complementary subunit interface has a positive electrostatic potential to interact with the partial negative charge of the lactam oxygen of strychnine.
Extended Data Figure 5 Sequence alignment of GlyRα3cryst with representative eukaryotic Cys-loop receptor family members.
Residue conservation is indicated by grey and black highlights. Site of N-linked glycosylation of GlyRα3cryst is indicated by the orange hexagon. Residues involved in binding of strychnine are indicated by green (the principal subunit) and cyan (the complementary subunit) dots. Red triangles above residues indicate mutations in GlyRα1 or GlyRβ that cause hyperekplexia. Signal peptides have been removed from all protein sequences. Secondary structure elements are denoted by cylinders (helices) and arrows (strands) above the alignment. The alignment was generated using ClustalW. Protein sequences are from the following entires: human GlyRα1 (Uniprot P23415), human GlyRβ (Uniprot 48167), GluClcryst (PDB accession number 4TNV), GABAAR-β3cryst (PDB accession number 4COF), and 5HT3A (PDB accession number 4PIR).
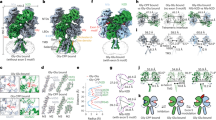
Extended Data Figure 6 Structural alignment of pLGICs.
a, Structures of GlyRα3cryst (green), apo-GluClα (slate), and glutamate- and ivermectin-bound GluCLα (violet) aligned using their ECDs. b, Close-up view of the loop C, rotated 45 ° clockwise along the horizontal axis (left to right) to optimize viewing. c, Close-up view of loops β6–β7 and β1–β2. d, Table showing regions of GlyRα3 used for structural alignments, as well as resulting number of overlapped Cα atoms and root mean squared deviation distances. PDB accession numbers are listed for each reference structure.
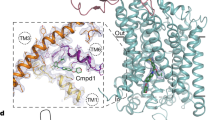
Extended Data Figure 7 Representative electron density for the ECD–TMD interface.
Final 2Fo – Fc electron density map around the region of the ECD–TMD interface is shown, contoured at 1.0σ. Protomer A is shown in pale green, protomer B in cyan, and protomer C in magenta. Important secondary structure elements and loops are noted.
Rights and permissions
About this article
Cite this article
Huang, X., Chen, H., Michelsen, K. et al. Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature 526, 277–280 (2015). https://doi.org/10.1038/nature14972
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14972
This article is cited by
-
AAV-glycine receptor α3 alleviates CFA-induced inflammatory pain by downregulating ERK phosphorylation and proinflammatory cytokine expression in SD rats
Molecular Medicine (2023)
-
Alkaline taste sensation through the alkaliphile chloride channel in Drosophila
Nature Metabolism (2023)
-
Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs
Nature Communications (2022)
-
Biosynthesis of strychnine
Nature (2022)
-
The glycine-containing dipeptide leucine-glycine raises accumbal dopamine levels in a subpopulation of rats presenting a lower endogenous dopamine tone
Journal of Neural Transmission (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.