Abstract
Protein aggregates are the hallmark of stressed and ageing cells, and characterize several pathophysiological states1,2. Healthy metazoan cells effectively eliminate intracellular protein aggregates3,4, indicating that efficient disaggregation and/or degradation mechanisms exist. However, metazoans lack the key heat-shock protein disaggregase HSP100 of non-metazoan HSP70-dependent protein disaggregation systems5,6, and the human HSP70 system alone, even with the crucial HSP110 nucleotide exchange factor, has poor disaggregation activity in vitro4,7. This unresolved conundrum is central to protein quality control biology. Here we show that synergic cooperation between complexed J-protein co-chaperones of classes A and B unleashes highly efficient protein disaggregation activity in human and nematode HSP70 systems. Metazoan mixed-class J-protein complexes are transient, involve complementary charged regions conserved in the J-domains and carboxy-terminal domains of each J-protein class, and are flexible with respect to subunit composition. Complex formation allows J-proteins to initiate transient higher order chaperone structures involving HSP70 and interacting nucleotide exchange factors. A network of cooperative class A and B J-protein interactions therefore provides the metazoan HSP70 machinery with powerful, flexible, and finely regulatable disaggregase activity and a further level of regulation crucial for cellular protein quality control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hipp, M. S., Park, S. H. & Hartl, F. U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014)
Morimoto, R. I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 (2008)
Kirstein-Miles, J., Scior, A., Deuerling, E. & Morimoto, R. I. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 32, 1451–1468 (2013)
Rampelt, H. et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31, 4221–4235 (2012)
Goloubinoff, P., Mogk, A., Zvi, A. P., Tomoyasu, T. & Bukau, B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl Acad. Sci. USA 96, 13732–13737 (1999)
Parsell, D. A., Kowal, A. S., Singer, M. A. & Lindquist, S. Protein disaggregation mediated by heat-shock protein HSP104. Nature 372, 475–478 (1994)
Shorter, J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 6, e26319 (2011)
Mayer, M. P. & Bukau, B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 (2005)
Kampinga, H. H. & Craig, E. A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature Rev. Mol. Cell Biol. 11, 579–592 (2010)
Cyr, D. M. & Ramos, C. H. Specification of Hsp70 function by type I and type II HSP40. Subcell. Biochem. 78, 91–102 (2015)
Sahi, C. & Craig, E. A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl Acad. Sci. USA 104, 7163–7168 (2007)
Tzankov, S., Wong, M. J., Shi, K., Nassif, C. & Young, J. C. Functional divergence between co-chaperones of Hsc70. J. Biol. Chem. 283, 27100–27109 (2008)
Rauch, J. N. & Gestwicki, J. E. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro . J. Biol. Chem. 289, 1402–1414 (2014)
Lu, Z. & Cyr, D. M. Protein folding activity of HSP70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273, 27824–27830 (1998)
Weibezahn, J. et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665 (2004)
Ramos, C. H., Oliveira, C. L., Fan, C. Y., Torriani, I. L. & Cyr, D. M. Conserved central domains control the quaternary structure of type I and type II HSP40 molecular chaperones. J. Mol. Biol. 383, 155–166 (2008)
Borges, J. C., Fischer, H., Craievich, A. F. & Ramos, C. H. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 280, 13671–13681 (2005)
Tsai, J. & Douglas, M. G. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 271, 9347–9354 (1996)
Suh, W. C., Lu, C. Z. & Gross, C. A. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem. 274, 30534–30539 (1999)
Genevaux, P., Schwager, F., Georgopoulos, C. & Kelley, W. L. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics 162, 1045–1053 (2002)
De Los Rios, P., Ben-Zvi, A., Slutsky, O., Azem, A. & Goloubinoff, P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl Acad. Sci. USA 103, 6166–6171 (2006)
Caughey, B. & Lansbury, P. T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 (2003)
Cheetham, M. E. & Caplan, A. J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28–36 (1998)
Li, J., Qian, X. & Sha, B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11, 1475–1483 (2003)
Lu, Z. & Cyr, D. M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273, 5970–5978 (1998)
Andréasson, C., Fiaux, J., Rampelt, H., Mayer, M. P. & Bukau, B. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283, 8877–8884 (2008)
Cashikar, A. G., Duennwald, M. & Lindquist, S. L. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J. Biol. Chem. 280, 23869–23875 (2005)
Haslbeck, M., Miess, A., Stromer, T., Walter, S. & Buchner, J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 280, 23861–23868 (2005)
Carra, S. et al. Different anti-aggregation and pro-degradative functions of the members of the mammalian sHSP family in neurological disorders. Phil. Trans. R. Soc. Lond. B 368, 20110409 (2013)
Vos, M. J. et al. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum. Mol. Genet. 19, 4677–4693 (2010)
Skouri-Panet, F., Michiel, M., Ferard, C., Duprat, E. & Finet, S. Structural and functional specificity of small heat shock protein HspB1 and HspB4, two cellular partners of HspB5: role of the in vitro hetero-complex formation in chaperone activity. Biochimie 94, 975–984 (2012)
Peschek, J. et al. Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc. Natl Acad. Sci. USA 110, E3780–E3789 (2013)
Leitner, A. et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126 (2012)
Rinner, O. et al. Identification of cross-linked peptides from large sequence databases. Nature Methods 5, 315–318 (2008)
Walzthoeni, T. et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nature Methods 9, 901–903 (2012)
Adams, S. R. et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124, 6063–6076 (2002)
Spagnuolo, C. C., Vermeij, R. J. & Jares-Erijman, E. A. Improved photostable FRET-competent biarsenical-tetracysteine probes based on fluorinated fluoresceins. J. Am. Chem. Soc. 128, 12040–12041 (2006)
Dixit, A., Ray, K., Lakowicz, J. R. & Black, L. W. Dynamics of the T4 bacteriophage DNA packasome motor: endonuclease VII resolvase release of arrested Y-DNA substrates. J. Biol. Chem. 286, 18878–18889 (2011)
Guex, N., Peitsch, M. C. & Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 (suppl. 1). S162–S173 (2009)
Kopp, J. & Schwede, T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 32, D230–D234 (2004)
Kiefer, F., Arnold, K., Kunzli, M., Bordoli, L. & Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 (2009)
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006)
Suzuki, H. et al. Peptide-binding sites as revealed by the crystal structures of the human Hsp40 Hdj1 C-terminal domain in complex with the octapeptide from human Hsp70. Biochemistry 49, 8577–8584 (2010)
Qian, Y. Q., Patel, D., Hartl, F. U. & McColl, D. J. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J. Mol. Biol. 260, 224–235 (1996)
Martinez, M. et al. SDA7: a modular and parallel implementation of the simulation of diffusional association software. J. Comput. Chem. 36, 1631–1645 (2015)
Gabdoulline, R. R. & Wade, R. C. Simulation of the diffusional association of barnase and barstar. Biophys. J. 72, 1917–1929 (1997)
Vriend, G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8, 52–56 (1990)
Madura, J. D. et al. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics Program. Comp. Phys. Comm. 91, 57–95 (1995)
Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996)
Gabdoulline, R. R. & Wade, R. C. Effective charges for macromolecules in solvent. J. Phys. Chem. 100, 3868–3878 (1996)
Elcock, A. H., Gabdoulline, R. R., Wade, R. C. & McCammon, J. A. Computer simulation of protein-protein association kinetics: acetylcholinesterase-fasciculin. J. Mol. Biol. 291, 149–162 (1999)
Gabdoulline, R. R. & Wade, R. C. On the contributions of diffusion and thermal activation to electron transfer between Phormidium laminosum plastocyanin and cytochrome f: Brownian dynamics simulations with explicit modeling of nonpolar desolvation interactions and electron transfer events. J. Am. Chem. Soc. 131, 9230–9238 (2009)
Barends, T. R. et al. Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ. Acta Crystallogr. D 69, 1540–1552 (2013)
Dragovic, Z., Broadley, S. A., Shomura, Y., Bracher, A. & Hartl, F. U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 (2006)
Raviol, H., Sadlish, H., Rodriguez, F., Mayer, M. P. & Bukau, B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 (2006)
Chen, J., Walter, S., Horwich, A. L. & Smith, D. L. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nature Struct. Biol. 8, 721–728 (2001)
Linke, K., Wolfram, T., Bussemer, J. & Jakob, U. The roles of the two zinc binding sites in DnaJ. J. Biol. Chem. 278, 44457–44466 (2003)
Garimella, R. et al. Hsc70 contacts helix III of the J domain from polyomavirus T antigens: addressing a dilemma in the chaperone hypothesis of how they release E2F from pRb. Biochemistry 45, 6917–6929 (2006)
Greene, M. K. et al. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl Acad. Sci. USA 95, 6108–6113 (1998)
Acknowledgements
We thank A. Mogk for critical reading of the manuscript and S. Ungelenk for Hsp26. This work was funded by the Deutsche Forschungsgemeinschaft (SFB1036, BU617/19-1 to B.B.; EXC257, SFB740 to J.K.), Alexander von Humboldt Foundation Postdoctoral Fellowships (to N.B.N. and A.Sz.), National Institutes of Health (the NIGMS, NIA, NIMS), Ellison Medical Foundation and Daniel F. and Ada L. Rice Foundation (to R.I.M.), German Federal Ministry of Education and Research (BMBF) Virtual Liver Network and EU FEP Flagship Programme Human Brain Project (0315749, 604102 to R.C.W.), Klaus Tschira Foundation (to M.B., A.St. and R.C.W.), Sir Henry Wellcome Postdoctoral Fellowship (to F.S.), ETH Zurich and ERC advanced grant Proteomics v3.0 (233226 to R.A.).
Author information
Authors and Affiliations
Contributions
N.B.N. and B.B. conceived the study. N.B.N., J.K., A.Sz., M.B., A.St., F.S., D.L.G., R.C.W., M.P.M. and B.B. designed the experiments. N.B.N., J.K., A.Sz., M.B., A.St., F.S., K.A., X.G. and A.Sc. performed the experiments. N.B.N., J.K., A.Sz., M.B., A.St., F.S., R.A., R.C.W., R.I.M., D.L.G., M.P.M. and B.B. analysed the data. N.B.N., D.L.G., M.P.M. and B.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Characterization of protein disaggregation/refolding and refolding-only reactions.
a, HSP70–J-protein–HSP110 (HSPA8–J-protein–HSPH2) functional cycle. Concomitant interaction of HSP70 with a J-protein and substrate results in allosteric stimulation of ATP hydrolysis; this traps the substrate in HSP70 (ref. 8). Subsequent NEF (for example, HSP110) promoted ADP dissociation from HSP70, then allows ATP rebinding, which triggers substrate release to complete the cycle54,55. b, Scheme for in vitro disaggregation/refolding and refolding-only reactions. The aggregates used in disaggregation/refolding assays are preformed by heating luciferase with yeast small heat-shock protein (sHSP) Hsp26 (ref. 4), which is known to co-aggregate with misfolded proteins in vivo27,28 (see Methods for detailed description). If HSP70, J-protein and HSP110 are instead heated together with substrate and Hsp26, luciferase is denatured into a more easily refoldable, inactive and largely monomeric substrate form used in refolding-only assays. c, SEC profiles of aggregated 3H-labelled luciferase (black; size range 200 kDa to ≥5,000 kDa representing ∼2 to >50 aggregated luciferase molecules) and monomeric native luciferase (red; size ∼63 kDa). Arrows indicate elution size (kDa). Inset, activity of loaded material. d, SEC profile of partially denatured and largely monomeric luciferase (starting material for refolding-only reactions). Inset, activity of loaded material. e, Chaperone nomenclature. f, Disaggregation and reactivation of preformed luciferase aggregates using human HSP70–HSP110 with human J-proteins JA2, green; JB1, blue; JA2+JB1, magenta or no J-protein, black. Under limiting chaperone (HSP70/HSP110) and increasing J-protein concentrations (A, solid or B, dashed) (n = 3). Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
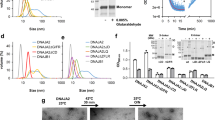
Extended Data Figure 2 Effects of mixed-class J-proteins on disaggregation/refolding and refolding-only activity of the HSP70 system.
a, Disaggregation/refolding of aggregated luciferase compared for human class A (JA1 and JA2) and class B (JB1 and JB4) J-proteins (n = 3). b, Luciferase refolding-only compared for JA1, JA2, JB1 and JB4 (n = 3). c, Reactivation of heat-aggregated luciferase with nematode HSP70 machinery, using reduced substrate:HSP70 ratio of 1:20, containing DNJ-12 (A), DNJ-13 (B) or DNJ-12+DNJ-13 (A+B) (n = 2). d, Disaggregation/refolding of luciferase using human HSP70 and HSP110 combined with nematode J-proteins (n = 3). e, Reactivation of luciferase showing optimal JA2:JB1 ratio for disaggregation/refolding (n = 2). f, Initial disaggregation/refolding rates for e. g, Final yields of refolded luciferase (120 min) for e. Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended Data Figure 3 Disaggregation synergy is independent of sHSP incorporation, NEF, substrate and aggregate character, and is not explained by sequential J-protein class activity.
a, Disaggregation/refolding reaction for luciferase aggregates without incorporating sHSP Hsp26 (n = 3). b, Reactivation without NEF (HSPH2) (n = 3). c, Reactivation of α-glucosidase aggregates (n = 3). d, Reactivation of preformed MDH aggregates in the presence of GroEL plus the GroES protein foldase system (GroELS) (n = 2). GroELS is required for efficient MDH refolding56. GroELS alone is in black. JB1:JA2 denotes the stoichiomety of each reaction. e, Disaggregation/refolding of stringent aggregates (≥5,000 kDa) formed using 2 μM luciferase (n = 3). f, Disaggregation/refolding of aggregated luciferase at reduced substrate:HSP70 ratio (luciferase:HSP70:J-protein:HSP110 = 1:7.5:3.8:0.4) (n = 3). The aggregated luciferase concentration is 100 nM. g, h, Holdase function of J-proteins (class A (g) and class B (h)) during luciferase aggregation at 42 °C, shown by decreased light scattering. Concentrations: 1× luciferase; 4× J-protein; 4× BSA (control) (n = 2). i, Reactivation with sequential JA2 and JB1 addition. J-protein added at t = 0 min (black graph legends); J-protein added after 30 or 60 min (red graph legends and arrows) (n = 2). Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended Data Figure 4 Stoichiometry of class A and B J-proteins determines the range of aggregate sizes resolved.
a, The GroEL trap (GroELD87K) facilitates the capture of 3H-luciferase monomers liberated by protein disaggregation before the refolding step. b, Refolding of disaggregated 3H-luciferase monomers (40 min) in the absence (solid bars) and presence of GroEL trap (open bars). c, SEC profile after disaggregation/refolding of aggregated tritiated α-glucosidase (60 min) with either J-protein class alone (green (A) or blue (B)) or J-proteins combined (magenta). Control reaction without chaperones (black). Elution fractions F1–F4 (red lines). Table shows size distribution of aggregates in each fraction; F1 luciferase aggregates ≥4,000 kDa; F2, aggregates ∼400–4,000 kDa; F3, aggregates ∼150–400 kDa, F4 disaggregated monomers (∼68 kDa). d, Quantification of SEC profile measuring disaggregation of tritiated α-glucosidase from aggregates (F1–F3) from c, also showing concomitant accumulation of disaggregated monomer (F4) from c (n = 3). e, ATP depletion by apyrase abrogates disaggregation. f, Quantification of SEC profile measuring disaggregation of tritiated luciferase from aggregates (F1–F3) with concomitant accumulation of disaggregated monomer (F4), using the HSP70–HSP110 system with JA2 or JB1 alone, or with JA2 plus JB1. Stoichiometry range used for JA2:JB1, 1:1 to 4:1 to 1:4. Specifically, 0.2 JB1:0.8 JA2 (orange); 0.2 JA2:0.8 JB1 (red). Solid colours denote 40-min reaction time; hash denotes 120 min. Control reaction without chaperones (black). Two-tailed t-test, *P < 0.05, **P < 0.01 (n = 3). Data are mean ± s.e.m. Precise concentrations are shown in Extended Data Table 1.
Extended Data Figure 5 JA2 and JB1 form homodimers and interact transiently.
a, Identified JA2 and JB1 inter-molecular cross-links; ‘Id’, amino acid sequence of peptides showing cross-linked lysines (K, orange). Protein 1 and 2 denote source proteins for cross-linked peptides; position 1 and 2 denote positions of cross-linked lysines within proteins; deltaS is the delta score for each crosslink; cut-off = 0.9. ld-Score is the linear discriminant score. b, Representative mass spectrometry spectra for inter-molecular JA2 and JB1 cross-links. Common peaks, green; cross-linked, red; matched peaks, diamonds (no peaks above 1,100 m/z detected). c, SEC profiles of 3H-labelled JA2 dimer (green cartoon) and 3H-labelled JB1 dimer (blue cartoon) mixed with unlabelled J-protein from the other class. Precise concentrations are shown in Extended Data Table 1.
Extended Data Figure 6 Electrostatic interactions between J-domain and CTD predominate in JA2 and JB1 complexes.
a, FRET efficiencies for JD–CTD and CTD–CTD interactions with 0–0.2% Tween20 titration. Percentage efficiency is relative to untreated (0% Tween20) samples. Donor quenching (black); acceptor fluorescence (red); below, fluorophore positions in J-protein protomers (JA2, green; JB1, blue). N-termini of JDJA2 and JDJB1 labelled with acceptor fluorophore ReAsH. CTDJA2 and CTDJB1 labelled with donor fluorophores FlAsH and Alexa Fluor 488 at residues 241 and 278, respectively. b, Disaggregation/refolding of preformed luciferase by JA2 and/or JB1 with increasing amounts of Tween20 (n = 2). c, FRET efficiencies for JA2 and JB1 interactions at increasing salt concentrations. d, Disaggregation/refolding of preformed luciferase aggregates by JA2 and JB1 with increasing salt concentrations; control, 50 mM salt, no chaperones (n = 2). e, Luciferase disaggregation/refolding in the presence of excess J-domain fragments carrying JD-QPNJA2/JB1 mutation of the HPD motif (n = 3). f, g, FRET between class A and B J-proteins. f, Competition with unlabelled full-length wild-type J-protein (FL); unlabelled competitor is 1–10× acceptor; (−), no competitor. g, Competition with unlabelled isolated JDJA2 and JDJB1. Data are mean ± s.e.m., average of at least two experiments for FRET experiments. Precise concentrations are shown in Extended Data Table 1.
Extended Data Figure 7 In silico prediction of JD–CTD interactions between class A and B J-proteins and in vitro evidence that physical interactions between J-proteins do not overlap J-protein substrate binding sites.
a, Preferred positions of the centres of geometry (CoG) of J-domains (y axis, JA1, JA2, JB1 and JB4) around CTD dimers (x axis, class A, green, class B, blue) obtained from molecular docking simulations. JDJA1/JB1, wireframe meshes; JDJA2/JB4, brown contours, each contoured at the isovalue given in the top left of each image. The higher scores for class A CTDs indicate greater specificity of the complexes formed with J-domains; the lower scores for class B CTDs indicate much less specific interactions. Lysines in inter- and intra-J-protein JA2–JB1 cross-links, orange spheres. b, Properties of the docking arrangements obtained after clustering. Total number of clusters per simulation, denominator; number of selected clusters (corresponding to 90% of all docked complexes), numerator, bold. In parentheses, the range of average energy values (in units of kT) for the selected clusters. Lower energy values indicate more favourable binding; fewer clusters indicate a more defined binding mode (see Methods). JDJA2 docking to CTDJB1 is much weaker and less specific than JDJB1 docking to CTDJA1, but docking arrangements compatible with cross-linking results still obtain (Fig. 2d). c, Competition of isolated JDJA2 fragments against JA2 holdase function in luciferase aggregation at 42 °C (n = 2). d, Competition of isolated JDJB1 fragments against JA2 holdase function (n = 2). Luciferase, 1×; JA2, 4×; isolated J-domain fragments, 20× (red; 5-fold excess over JA2), or 40× (orange; 10-fold excess over JA2). Light scattering measured at 600 nm. Precise concentrations are shown in Extended Data Table 1.
Extended Data Figure 8 Possible configurations of the JA2–JB1 mixed-class complex.
a, Compact configuration. b, Open configuration. Configurations were derived from computational docking, using constraints from experimental FRET and cross-linking data (Fig. 2a–d and Extended Data Fig. 5a). Each configuration is shown from two views (left and right) rotated by 135 degrees with respect to each other and in ribbon (top) and molecular surface (bottom) representations. In both cases J-domains of JA2 dock onto the CTD dimer of JB1, and similarly J-domains of JB1 dock to the CTD dimer of JA2. Both CTDJB1 protomers are within cross-linking distance of CTDJA2. Unstructured glycine/phenylalanine (G/F)-rich flexible regions connecting J-domains and CTDs shown by dark blue (JB1) or green (JA2) dashed lines. Residues at FRET fluorophore sites are shown in space-filling representation (red on JA2, magenta on JB1). Inter-molecular crosslinking lysine pairs (gold and cyan, space-filling) are connected by dotted lines. Bottom left within a: molecular surface representation of compact configuration of the JA2–JB1 complex, showing substrate binding sites from crystallographic24 (yellow) and biochemical57 (orange, cyan) data. HPD motif, red. Residues implicated in JD–HSP70 interactions19,58,59 (dark teal and dark green on JDJA2; purple and dark blue on JDJB1). Bottom right within a: rotated image. Table shows fluorophore separation distances; calculated percentage FRET efficiencies in parentheses. a, Both CTDJB1 protomers are within cross-linking distance of CTDJA2. b, As in a, but with only a single CTDJB1 protomer within cross-linking distance to CTDJA2; one JDJA2 docks onto CTDJB1, the other JDJA2 is free. Similarly, one JDJB1 docks onto CTDJA2, the other JDJB1 docks onto its own CTD, consistent with SAXS-determination of class B J-proteins16,17. Model of JB1 (blue) based on the crystal structure of CTD and NMR structure of J-domain. Homology model of JA2 (green) based on the crystal structure of Ydj1 (see Methods).
Rights and permissions
About this article
Cite this article
Nillegoda, N., Kirstein, J., Szlachcic, A. et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251 (2015). https://doi.org/10.1038/nature14884
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14884
This article is cited by
-
The generation of detergent-insoluble clipped fragments from an ERAD substrate in mammalian cells
Scientific Reports (2023)
-
HSPA8 acts as an amyloidase to suppress necroptosis by inhibiting and reversing functional amyloid formation
Cell Research (2023)
-
Stress-induced protein disaggregation in the endoplasmic reticulum catalysed by BiP
Nature Communications (2022)
-
Identification of a HTT-specific binding motif in DNAJB1 essential for suppression and disaggregation of HTT
Nature Communications (2022)
-
Picturing protein disaggregation
Nature Chemical Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.