Abstract
Biomolecular self-assemblies are of great interest to nanotechnologists because of their functional versatility and their biocompatibility1. Over the past decade, sophisticated single-component nanostructures composed exclusively of nucleic acids2,3,4,5, peptides6,7,8 and proteins9,10,11,12,13,14,15 have been reported, and these nanostructures have been used in a wide range of applications, from drug delivery16 to molecular computing17. Despite these successes, the development of hybrid co-assemblies of nucleic acids and proteins has remained elusive. Here we use computational protein design to create a protein–DNA co-assembling nanomaterial whose assembly is driven via non-covalent interactions. To achieve this, a homodimerization interface is engineered onto the Drosophila Engrailed homeodomain (ENH), allowing the dimerized protein complex to bind to two double-stranded DNA (dsDNA) molecules. By varying the arrangement of protein-binding sites on the dsDNA, an irregular bulk nanoparticle or a nanowire with single-molecule width can be spontaneously formed by mixing the protein and dsDNA building blocks. We characterize the protein–DNA nanowire using fluorescence microscopy, atomic force microscopy and X-ray crystallography, confirming that the nanowire is formed via the proposed mechanism. This work lays the foundation for the development of new classes of protein–DNA hybrid materials. Further applications can be explored by incorporating DNA origami, DNA aptamers and/or peptide epitopes into the protein–DNA framework presented here.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sarikaya, M., Tamerler, C., Jen, A. K. Y., Schulten, K. & Baneyx, F. Molecular biomimetics: nanotechnology through biology. Nature Mater. 2, 577–585 (2003)
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991)
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003)
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006)
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009)
Zhang, S., Holmes, T., Lockshin, C. & Rich, A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl Acad. Sci. USA 90, 3334–3338 (1993)
Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nature Biotechnol. 21, 1171–1178 (2003)
Fletcher, J. M. et al. Self-assembling cages from coiled-coil peptide modules. Science 340, 595–599 (2013)
Brodin, J. D. et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nature Chem. 4, 375–382 (2012)
Lai, Y. T., Cascio, D. & Yeates, T. O. Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129 (2012)
King, N. P. et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012)
Lanci, C. J. et al. Computational design of a protein crystal. Proc. Natl Acad. Sci. USA 109, 7304–7309 (2012)
King, N. P. et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014)
Huang, P. S. et al. High thermodynamic stability of parametrically designed helical bundles. Science 346, 481–485 (2014)
Thomson, A. R. et al. Computational design of water-soluble α-helical barrels. Science 346, 485–488 (2014)
Chang, M., Yang, C. S. & Huang, D. M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano 5, 6156–6163 (2011)
Amir, Y. et al. Universal computing by DNA origami robots in a living animal. Nature Nanotechnol. 9, 353–357 (2014)
Sanchez, C., Julian, B., Belleville, P. & Popall, M. Applications of hybrid organic–inorganic nanocomposites. J. Mater. Chem. 15, 3559–3592 (2005)
Grigoryan, G. et al. Computational design of virus-like protein assemblies on carbon nanotube surfaces. Science 332, 1071–1076 (2011)
Niemeyer, C. M., Koehler, J. & Wuerdemann, C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H:FMN oxidoreductase and luciferase. ChemBioChem 3, 242–245 (2002)
Sano, T., Smith, C. L. & Cantor, C. R. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 258, 120–122 (1992)
Wilner, O. I., Shimron, S., Weizmann, Y., Wang, Z. G. & Willner, I. Self-assembly of enzymes on DNA scaffolds: en route to biocatalytic cascades and the synthesis of metallic nanowires. Nano Lett. 9, 2040–2043 (2009)
Delebecque, C. J., Lindner, A. B., Silver, P. A. & Aldaye, F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474 (2011)
Draganescu, A. & Tullius, T. D. The DNA binding specificity of engrailed homeodomain. J. Mol. Biol. 276, 529–536 (1998)
Guerrero, L., Smart, O. S., Woolley, G. A. & Allemann, R. K. Photocontrol of DNA binding specificity of a miniature engrailed homeodomain. J. Am. Chem. Soc. 127, 15624–15629 (2005)
Marshall, S. A., Morgan, C. S. & Mayo, S. L. Electrostatics significantly affect the stability of designed homeodomain variants. J. Mol. Biol. 316, 189–199 (2002)
Shah, P. S. et al. Full-sequence computational design and solution structure of a thermostable protein variant. J. Mol. Biol. 372, 1–6 (2007)
Mou, Y., Huang, P.-S., Hsu, F.-C., Huang, S. -J. & Mayo, S. L. Computational design and experimental verification of a symmetric protein homodimer. Proc. Natl Acad. Sci. USA http://dx.doi.org/10.1073/pnas.1505072112 (2015)
Mou, Y., Huang, P.-S., Thomas, L. M. & Mayo, S. L. Using molecular dynamics simulations as an aid in the prediction of domain swapping of computationally designed protein variants. J. Mol. Biol 427, 2697–2706 (2015)
Whaley, S. R., English, D. S., Hu, E. L., Barbara, P. F. & Belcher, A. M. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 405, 665–668 (2000)
Clarke, N. D., Kissinger, C. R., Desjarlais, J., Gilliland, G. L. & Pabo, C. O. Structural studies of the engrailed homeodomain. Protein Sci. 3, 1779–1787 (1994)
Huang, P.-S., Love, J. J. & Mayo, S. L. Adaptation of a fast Fourier transform-based docking algorithm for protein design. J. Comput. Chem. 26, 1222–1232 (2005)
O’Shea, E. K., Klemm, J. D., Kim, P. S. & Alber, T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 (1991)
Das, R. & Baker, D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 77, 363–382 (2008)
Allen, B. D. & Mayo, S. L. Dramatic performance enhancements for the FASTER optimization algorithm. J. Comput. Chem. 27, 1071–1075 (2006)
Dunbrack, R. L. Jr & Karplus, M. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J. Mol. Biol. 230, 543–574 (1993)
Stemmer, W. P., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 (1995)
Fraenkel, E., Rould, M. A., Chambers, K. A. & Pabo, C. O. Engrailed homeodomain-DNA complex at 2.2 Å resolution: a detailed view of the interface and comparison with other engrailed structures. J. Mol. Biol. 284, 351–361 (1998)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
This study was supported by the Defense Advanced Research Projects Agency Protein Design Processes Program, a National Security Science and Engineering Faculty Fellowship (NSSEFF N00244-09-1-0011, N00244-09-1-0082), and the Gordon and Betty Moore Foundation through grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative. We would like to acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech, and the Department of Energy and National Institutes of Health for supporting the Stanford Synchrotron Radiation Lightsource. We thank J. Kaiser, J. Hoy and P. Nikolovski at the Caltech Molecular Observatory for assistance in crystal screening and crystallographic data collection. Y.M. thanks L.-C. Ho for her encouragement and literature research in the crystallographic work. Y.M. thanks T. J. Zwang for assistance with AFM measurements. Y.M. thanks X. Zhang and S. Yan for the useful discussion. We are grateful to J. Kostecki and M. Ary for assistance with the manuscript.
Author information
Authors and Affiliations
Contributions
Y.M. designed and performed the experiments. Y.M. and J.-Y.Y. performed the optical microscope experiments. All authors wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Design model of irregular bulk protein–DNA nanoparticle.
a, Four consecutive ENH-binding sites that each face in a different direction are engineered onto a dsDNA fragment. This dsDNA building block allows the protein–DNA assembly to occur in all three dimensions. Note that in this particular design, two neighbouring binding sites may not be simultaneously occupied due to steric hindrance. b, Cartoon illustrating an irregular shaped nanoparticle formed by co-assembly of dualENH and the dsDNA shown in a. The DNA-binding domains of dualENH are shown as blue triangles, and the homodimerization domains are shown as green squares.
Extended Data Figure 2 Circular dichroism spectroscopy of dualENH.
a, Circular dichroism spectrum of dualENH at room temperature. Solid line: before thermal denaturation; dashed line: after thermal denaturation. The overlapping of the two curves indicates that dualENH folds reversibly. b, Thermal denaturation curve measured at 222 nm. Circles: experimental data; line: curve fit with two-state transition model. The melting temperature of dualENH was determined to be 59 °C.
Extended Data Figure 3 Biophysical characterization of dualENH.
a, Size-exclusion chromatography of dualENH with three different loading concentrations: solid line, 650 μM; dashed line, 80 μM; dotted line, 5 μM. The highest signals were normalized to 100 for all curves. b, c(S) model fit from a sedimentation velocity experiment of 40 μM dualENH. The major peak around S = 1.9 corresponds to a molecular weight of 18.3 kDa, which is about twice that of monomeric dualENH (8.7 kDa). The spike at the left (S < 0.5) may be due to impurities or artefacts from model fitting. c, Raw data and fitting residuals for the sedimentation velocity experiment in b. A total of 378 curves were used for fitting, but for visual clarity only one-fifth of the curves are shown. The top graph shows the raw data (dots) and the fitting curves; the bottom shows the residuals between the experimental data and the fit. The square root of variance of the fit is 0.00669. d, Fluorescence polarization experiment. Two dsDNA sequences labelled with fluorescein were used as probes to assay dualENH–DNA binding. Probe 1: 20-nucleotide dsDNA with the binding motif TAATTA; probe 2: same sequence as probe 1 but with a single-nucleotide mutation to the binding motif (TA[C]TTA). The concentration of dualENH was varied, while the concentration of the three probes remained constant (25 nM). Data are shown as mean ± standard error of the mean (s.e.m.) for three replicates. e, FRET experiment showing that dualENH brings two dsDNA fragments within Förster distance. Fifteen-nucleotide dsDNA (TAA)5 were labelled with either Cy3 or Cy5 to serve as the FRET donor or acceptor. Grey line: 400 nM Cy3-(TAA)5 plus 600 nM Cy5-(TAA)5; black line: 400 nM Cy3-(TAA)5 plus 600 nM Cy5-(TAA)5 plus 4 μM dualENH. f, Two control experiments for the FRET experiment in e. Black line: 400 nM Cy3-(TAA)5; black dashed line: 400 nM Cy3-(TAA)5 plus 4 μM dualENH; grey line: 600 nM Cy5-(TAA)5; grey dashed line: 600 nM Cy5-(TAA)5 plus 4 μM dualENH.
Extended Data Figure 4 Fluorescence polarization experiments with dualENH and wild-type ENH.
a, Fluorescence polarization experiment with probe 1, which contains the binding motif TAATTA of ENH. The concentration of dualENH or wild-type ENH was varied, while the concentration of probe 1 remained constant (25 nM). Both dualENH and wild-type ENH show saturated binding when the protein concentration is at or above 100 nM. Data are shown as mean ± s.e.m. for three replicates. b, Same experiments as a, except that a probe without any TAATNN binding motif was used. Note that probe 1 used in a has a lower fluorescence intensity and polarization than the probe used in b, probably due to partial quenching by a guanine nucleotide on the strand opposite the fluorescein label. Data are shown as mean ± s.e.m. for three replicates.
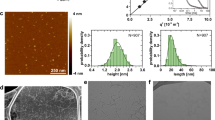
Extended Data Figure 5 Microscope imaging experiments.
a, The size distribution of the irregular protein–DNA particles formed by 5 µM dualENH mixed with 2 µM Cy3-(TAA)5. b, Bright-field microscopy image of 5 µM dualENH mixed with 2 µM Cy3-(TAA)5. A dust particle (top left) is evident, indicating that the focal plane is correct. c, Fluorescence microscopy image of 2 µM Cy3-(TAA)5 alone. d, Fluorescence microscopy image of particles formed with 500 nM dualENH mixed with 200 nM Cy3-(TAA)5. e, Fluorescence microscopy image of particles formed with 200 nM dualENH mixed with 100 nM Cy3-(TAA)5. f, Fluorescence microscopy image of particle inhibition experiments. A small amount (5 nM) of single-binding-site dsDNA (containing only one TAATTA motif) was pre-mixed with 500 nM dualENH, then 200 nM Cy3-(TAA)5 was added. The illumination/camera sensitivity was enhanced to confirm that particle formation is nearly completely absent under these conditions.
Extended Data Figure 6 Co-crystal structure of the protein–DNA complex.
a, Structures in the asymmetric unit cell are shown in colour, and the end-to-end packing of neighbouring DNA molecules and their bound proteins are shown in grey. b, c, The dualENH homodimer observed in the co-crystal structure (green or cyan) is superimposed with the design model (grey). The backbone r.m.s.d. compared to the design model is 3.8 Å (green) and 3.9 Å (cyan), respectively. When only one subunit is aligned between the more predominant configuration (green) and the design model, the angular displacement between the other subunits is ∼45° with about 3 Å translational displacement. The less predominant configuration has a lower angular displacement, ∼20°, but a larger translational displacement, ∼8 Å. The calculated energies for the design model and the two crystallographic dimers are −155.2, −140.3 (green) and −131.2 (cyan) Rosetta energy units, respectively.
Extended Data Figure 7 dualENH–DNA binding and nanostructure formation are inhibited at high salt concentrations.
a, Fluorescence polarization experiments of dualENH and probe 1 at various NaCl concentrations. Probe 1 and dualENH were mixed in buffers with different NaCl concentrations and fluorescence polarization values were recorded. dualENH–DNA binding dropped greatly from 100 mM to 150 mM NaCl, and was completely absent at 300 mM NaCl. Data are shown as mean ± s.e.m. for three replicates. b, Fluorescence microscopy image of particle experiment at 150 mM salt concentration. The sample was prepared by mixing 500 nM dualENH and 200 nM Cy3-(TAA)5 in 20 mM Tris-HCl buffer with 150 mM NaCl. The illumination/camera sensitivity was enhanced to confirm that particle formation is nearly completely absent under these conditions.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion. (PDF 132 kb)
Rights and permissions
About this article
Cite this article
Mou, Y., Yu, JY., Wannier, T. et al. Computational design of co-assembling protein–DNA nanowires. Nature 525, 230–233 (2015). https://doi.org/10.1038/nature14874
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14874
This article is cited by
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)
-
Agglomeration: when folded proteins clump together
Biophysical Reviews (2023)
-
Programmable Oligonucleotide-Peptide Complexes: Synthesis and Applications
Chemical Research in Chinese Universities (2022)
-
Hybrid gold/DNA nanowire circuit with sub-10 nm nanostructure arrays
Microsystems & Nanoengineering (2020)
-
Eukaryotic transcription factors can track and control their target genes using DNA antennas
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.