Abstract
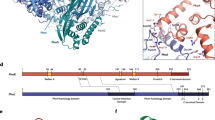
Phosphorus is required for all life and microorganisms can extract it from their environment through several metabolic pathways. When phosphate is in limited supply, some bacteria are able to use phosphonate compounds, which require specialized enzymatic machinery to break the stable carbon–phosphorus (C–P) bond. Despite its importance, the details of how this machinery catabolizes phosphonates remain unknown. Here we determine the crystal structure of the 240-kilodalton Escherichia coli C–P lyase core complex (PhnG–PhnH–PhnI–PhnJ; PhnGHIJ), and show that it is a two-fold symmetric hetero-octamer comprising an intertwined network of subunits with unexpected self-homologies. It contains two potential active sites that probably couple phosphonate compounds to ATP and subsequently hydrolyse the C–P bond. We map the binding site of PhnK on the complex using electron microscopy, and show that it binds to a conserved insertion domain of PhnJ. Our results provide a structural basis for understanding microbial phosphonate breakdown.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession code 4XB6. The EM density map has been deposited in the Electron Microscopy Data Bank (EMDB) with accession code EMD-3033.
References
Karl, D. M. Aquatic ecology: Phosphorus, the staff of life. Nature 406, 31–33 (2000)
Hsieh, Y. J. & Wanner, B. L. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203 (2010)
Hove-Jensen, B., Zechel, D. L. & Jochimsen, B. Utilization of glyphosate as phosphate source: biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol. Mol. Biol. Rev. 78, 176–197 (2014)
Imazu, K. et al. Enhanced utilization of phosphonate and phosphite by Klebsiella aerogenes . Appl. Environ. Microbiol. 64, 3754–3758 (1998)
Quinn, J. P. Carbon phosphorus lyase activity — a novel mechanism of bacterial-resistance to the phosphonic acid antibiotics. Lett. Appl. Microbiol. 8, 113–116 (1989)
Schowanek, D. & Verstraete, W. Phosphonate utilization by bacterial cultures and enrichments from environmental samples. Appl. Environ. Microbiol. 56, 895–903 (1990)
Wackett, L. P., Shames, S. L., Venditti, C. P. & Walsh, C. T. Bacterial carbon-phosphorus lyase: products, rates, and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 169, 710–717 (1987)
White, A. K. & Metcalf, W. W. Microbial metabolism of reduced phosphorus compounds. Annu. Rev. Microbiol. 61, 379–400 (2007)
Chen, C. M., Ye, Q. Z., Zhu, Z. M., Wanner, B. L. & Walsh, C. T. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B . J. Biol. Chem. 265, 4461–4471 (1990)
Hove-Jensen, B., McSorley, F. R. & Zechel, D. L. Physiological role of phnP-specified phosphoribosyl cyclic phosphodiesterase in catabolism of organophosphonic acids by the carbon-phosphorus lyase pathway. J. Am. Chem. Soc. 133, 3617–3624 (2011)
Hove-Jensen, B., Rosenkrantz, T. J., Haldimann, A. & Wanner, B. L. Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): dual role in phosphonate degradation and NAD biosynthesis pathways. J. Bacteriol. 185, 2793–2801 (2003)
Metcalf, W. W. & Wanner, B. L. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli . Gene 129, 27–32 (1993)
Rizk, S. S., Cuneo, M. J. & Hellinga, H. W. Identification of cognate ligands for the Escherichia coli phnD protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Sci. 15, 1745–1751 (2006)
Jochimsen, B. et al. Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway. Proc. Natl Acad. Sci. USA 108, 11393–11398 (2011)
Ren, Z. et al. Subunit interactions within the carbon-phosphorus lyase complex from Escherichia coli . Biochemistry 54, 3400–3411 (2015)
Frost, J. W., Loo, S., Cordeiro, M. L. & Li, D. Radical-based dephosphorylation and organophosphonate biodegradation. J. Am. Chem. Soc. 109, 2166–2171 (1987)
Kamat, S. S., Williams, H. J., Dangott, L. J., Chakrabarti, M. & Raushel, F. M. The catalytic mechanism for aerobic formation of methane by bacteria. Nature 497, 132–136 (2013)
Kamat, S. S., Williams, H. J. & Raushel, F. M. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature 480, 570–573 (2011)
Adams, M. A. et al. Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli . J. Bacteriol. 190, 1072–1083 (2008)
Shisler, K. A. & Broderick, J. B. Glycyl radical activating enzymes: structure, mechanism, and substrate interactions. Arch. Biochem. Biophys. 546, 64–71 (2014)
Booker, S. J. & Grove, T. L. Mechanistic and functional versatility of radical SAM enzymes. F1000 Biol. Rep. 2, 52 (2010)
Frey, P. A., Hegeman, A. D. & Ruzicka, F. J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 (2008)
Sofia, H. J., Chen, G., Hetzler, B. G., Reyes-Spindola, J. F. & Miller, N. E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 (2001)
Goldman, P. J. et al. X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl Acad. Sci. USA 110, 8519–8524 (2013)
McCall, K. A., Huang, C. & Fierke, C. A. Function and mechanism of zinc metalloenzymes. J. Nutr. 130, 1437S–1446S (2000)
Yakovleva, G. M., Kim, S. K. & Wanner, B. L. Phosphate-independent expression of the carbon-phosphorus lyase activity of Escherichia coli . Appl. Microbiol. Biotechnol. 49, 573–578 (1998)
Li, X. et al. Structure of the nucleotide-binding domain of a dipeptide ABC transporter reveals a novel iron-sulfur cluster-binding domain. Acta Crystallogr. D 69, 256–265 (2013)
Hollenstein, K., Dawson, R. J. & Locher, K. P. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 (2007)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
D’Arcy, A., Villard, F. & Marsh, M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D 63, 550–554 (2007)
Knäblein, J. et al. Ta6Br12 2+, a tool for phase determination of large biological assemblies by X-ray crystallography. J. Mol. Biol. 270, 1–7 (1997)
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Cohen, S. X. et al. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D 60, 2222–2229 (2004)
Thorn, A. & Sheldrick, G. M. Extending molecular-replacement solutions with SHELXE. Acta Crystallogr. D 69, 2251–2256 (2013)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Schrödinger, LLC. The PyMOL molecular graphics system, version 1.3r1. (2010)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Ermantraut, E., Wohlfart, K. & Tichelaar, W. Perforated support foils with pre-defined hole size, shape and arrangement. Ultramicroscopy 74, 75–81 (1998)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012)
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nature Methods 9, 853–854 (2012)
Russo, C. J. & Passmore, L. A. Robust evaluation of 3D electron cryomicroscopy data using tilt-pairs. J. Struct. Biol. 187, 112–118 (2014)
Neidhardt, F. C., Bloch, P. L. & Smith, D. F. Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974)
Acknowledgements
We are thankful to T. L. Sørensen at Diamond, T. Weinert at SLS as well as beamline staff at ESRF and MAX-Lab for help during X-ray data acquisition and S. Chen, C. G. Savva, J. Grimmett and T. Darling at the MRC-LMB for technical assistance with electron microscopy. This work was supported by the European Research Council grant no. 261151 (L.A.P.), MRC grant MC_U105192715 (L.A.P.), and the Danish National Research Foundation ‘Centre for mRNP biogenesis and metabolism’ (D.E.B.).
Author information
Authors and Affiliations
Contributions
P.S., L.A.P., B.H.J., B.J. and D.E.B. designed and P.S., L.B.V., C.J.R. and B.J. carried out the experiments. P.S., M.K. and D.E.B. determined the crystal and EM structures while C.J.R. and L.A.P. carried out final refinement of the EM structure as well as EM structure validation. P.S., M.K., C.J.R., L.A.P., B.H.J., B.J. and D.E.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
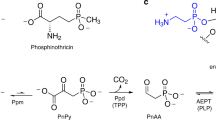
Extended Data Figure 1 The conversion of a phosphonate to 5-phosphoribosyl-α-1-diphosphate (PRPP).
PhnI supported by PhnG, PhnH and PhnL catalyses the transfer of the phosphonate moiety to the 1′ position of the ribose of ATP through displacement of adenine, generating a 5-triphosphoribosyl-α-1-phosphonate. Subsequent to the removal of pyrophosphate by PhnM yielding a 5-phosphoribosyl-α-1-phosphonate, PhnJ breaks the C–P bond of the ribose-coupled phosphonate liberating the alkyl moiety and generating 5-phosphoribosyl 1,2-cyclic phosphate. Finally, the combined activities of PhnP (a phosphoribosyl cyclic phosphodiesterase) and PhnN (a ribosyl bisphosphate phosphokinase) result in the formation of PRPP via ribose 1,5-bisphosphate.
Extended Data Figure 2 Representative examples of electron density.
a, The interface between PhnJ (blue, residues 45–52 and 104–110 including a bound sulfate ion) and PhnI (green, residues 321–341) showing 2Fo − Fc electron density contoured at 2.0 r.m.s.d. b, Close-up of the aromatic side chains in the central β-sheet of PhnJ (residues 118–126, 203–207 and 211–217), with the same contouring as a.
Extended Data Figure 3 Sequences of the proteins of the C–P lyase core complex with secondary structure.
Protein sequences are shown along with secondary structure assignment based on the crystal structure and colours as in Fig. 1. a, PhnG. The first α-helix is two residues longer in one of the two copies in the complex and indicated with a dashed box. The β-barrel domain is shown in yellow and orange colours. b, PhnH. The numbering of β-strands follows the convention from ref. 19. Helix names (A–E) used in that paper are shown in parentheses. β1 and β3 are not included in the figures in this paper as they only have two hydrogen bonds each. c, PhnI. The N-terminal domain is shown with sea green and the β-barrel domain with green and light-green colours. d, PhnJ. The central insertion domain is shown in purple and the C-terminal mini domain in a darker blue colour. Figure produced using SecSeq (D. E. Brodersen, unpublished, http://www.bioxray.au.dk/~deb/secseq).
Extended Data Figure 4 Cross alignments.
a, Alignment of the amino acid sequences of E. coli PhnH and PhnJ. Identical residues are shown in red and conserved functionality in green. Secondary structure colours correspond to Fig. 1 and conserved regions are shown in dashed boxes. For PhnJ, the positions of the CID and CMD are indicated with brackets as well as with colours. b, Alignment of PhnG and PhnI (only partial sequence). Conserved regions are shown in dashed boxes. Figure produced using SecSeq (D. E. Brodersen, unpublished, http://www.bioxray.au.dk/~deb/secseq).
Extended Data Figure 5 The structure and function of PhnJ.
a, Two perpendicular views of the PhnH–PhnJ heterodimer as observed within the C–P lyase core complex (blue and red, left) and aligned views of the PhnH2 homodimer from the isolated crystal structure (PDB ID: 2FSU; green, right)19. b, Surface view of the C–P lyase core complex with PhnG shown in yellow, PhnI in green, and PhnJ in blue. The position of SAM as modelled from an S-adenosyl methionine activase enzyme (PDB ID: 4K37)24 has been overlaid to visualize its putative placement in the pocket between PhnG and PhnJ. c, The C–P lyase core complex shown in surface representation with PhnJ and the CMD in cartoon, coloured by B factor to show flexibility (B = 25 Å2, blue, B = 45 Å2, magenta). The zinc and sulfate ions are shown with spheres.
Extended Data Figure 6 Interaction areas within the C–P lyase core complex.
a, Dimerization interface between halves of the (PhnGHIJ)2 complex. Colours as in Fig. 1. b, Interaction areas between the individual subunits within each dimer half. c, Two perpendicular views of the C–P lyase core complex shown in surface representation with colours as in Fig. 1c. d, Left, overview of the surface conservation of the C–P lyase core complex shown as a colour gradient from teal (variable) to burgundy (conserved) as indicated. Right, conservation at the interaction interfaces between the individual subunits of the complex.
Extended Data Figure 7 In vivo complementation of E. coli ΔphnHIJKLMNOP.
E. coli strain BW16711 (ΔphnHIJKLMNOP) complemented with a plasmid-borne copy of either wild-type (Wt) phnGHIJKLMNOP or variants thereof, including PhnJ(C272A), PhnJ(H108A), PhnI(H328A), or the PhnI(H328A;H333A) double variant. Growth is monitored on minimal plates with either no phosphorus source (a), 2-aminoethyl phosphonate (2-AEPn) (b), methyl phosphonate (MPn) (c), or phosphate ion (d). The data shown are representative of three independent experiments.
Extended Data Figure 8 PhnK sequence alignment.
Alignment of the protein sequence of E. coli PhnK with homologous proteins from a wide range of microorganisms. Conserved residues are shown on a teal background, and residues mentioned in the main text and the location of the ABC cassette consensus motifs (Walker A, Q motif, ABC motif, Walker B, D loop, and H motif) are indicated.
Extended Data Figure 9 Negative stain electron microscopy of PhnGHIJK.
a, Raw micrograph representative of 100 images collected. Scale bar is 500 Å. b, Selection of 2D reference-free class averages from a total of 300 classes showing the particle in various orientations. Each class is 172 Å wide. c, Fourier-shell correlation (FSC) of the final electron microscopy density map as a function of resolution (black line) with FSC = 0.143 at 16 Å. The red line shows the correlation between the crystal structure and the electron microscopy density, which has FSC = 0.5 at 28 Å. The inset figure shows an equal area projection plot for the electron microscopy tilt pair validation data set. The circle is an approximation of a 99% confidence interval that contains the representative direction (blue plus), includes the true tilt direction (red cross), and excludes the untitled direction (origin).
Extended Data Figure 10 Purification of the PhnGHIJ(ΔCID) and PhnGHIJ(ΔCID)K complexes.
a, Sequence alignment of the CID region of PhnJ with the corresponding part of the structural domain of PhnH, where strands 5 and 6 of the β-sheet are connected by a short Gly–Gly turn (green residues). In PhnJ(ΔCID), the CID domain spanning residues 130–171 (red residues) are replaced by a similar dipeptide turn, which should maintain the overall domain fold. b, SDS–PAGE gel showing purified C–P lyase complexes (PhnGHIJ and PhnGHIJK) both with (Wt) and without (ΔCID) the CID on PhnJ. PhnK is missing from the complex purified from the PhnGHIJ(ΔCID)K construct (red arrow). The data shown are representative of three independent purifications. c, Overview of the C–P lyase core complex structure docked in the PhnGHIJK electron microscopy density with the location of the PhnJ CID domains (purple) and PhnK (cyan cartoon) indicated.
Source data
Rights and permissions
About this article
Cite this article
Seweryn, P., Van, L., Kjeldgaard, M. et al. Structural insights into the bacterial carbon–phosphorus lyase machinery. Nature 525, 68–72 (2015). https://doi.org/10.1038/nature14683
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14683
This article is cited by
-
Structural remodelling of the carbon–phosphorus lyase machinery by a dual ABC ATPase
Nature Communications (2023)
-
PCycDB: a comprehensive and accurate database for fast analysis of phosphorus cycling genes
Microbiome (2022)
-
Rhodium-catalyzed selective direct arylation of phosphines with aryl bromides
Nature Communications (2022)
-
Peptidoglycan recycling mediated by an ABC transporter in the plant pathogen Agrobacterium tumefaciens
Nature Communications (2022)
-
Global and seasonal variation of marine phosphonate metabolism
The ISME Journal (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.