Abstract
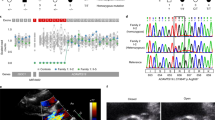
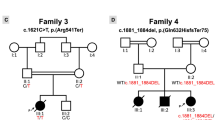
Mitral valve prolapse (MVP) is a common cardiac valve disease that affects nearly 1 in 40 individuals1,2,3. It can manifest as mitral regurgitation and is the leading indication for mitral valve surgery4,5. Despite a clear heritable component, the genetic aetiology leading to non-syndromic MVP has remained elusive. Four affected individuals from a large multigenerational family segregating non-syndromic MVP underwent capture sequencing of the linked interval on chromosome 11. We report a missense mutation in the DCHS1 gene, the human homologue of the Drosophila cell polarity gene dachsous (ds), that segregates with MVP in the family. Morpholino knockdown of the zebrafish homologue dachsous1b resulted in a cardiac atrioventricular canal defect that could be rescued by wild-type human DCHS1, but not by DCHS1 messenger RNA with the familial mutation. Further genetic studies identified two additional families in which a second deleterious DCHS1 mutation segregates with MVP. Both DCHS1 mutations reduce protein stability as demonstrated in zebrafish, cultured cells and, notably, in mitral valve interstitial cells (MVICs) obtained during mitral valve repair surgery of a proband. Dchs1+/− mice had prolapse of thickened mitral leaflets, which could be traced back to developmental errors in valve morphogenesis. DCHS1 deficiency in MVP patient MVICs, as well as in Dchs1+/− mouse MVICs, result in altered migration and cellular patterning, supporting these processes as aetiological underpinnings for the disease. Understanding the role of DCHS1 in mitral valve development and MVP pathogenesis holds potential for therapeutic insights for this very common disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Freed, L. A. et al. Mitral valve prolapse in the general population: the benign nature of echocardiographic features in the Framingham Heart Study. J. Am. Coll. Cardiol. 40, 1298–1304 (2002)
Avierinos, J. F. et al. Natural history of asymptomatic mitral valve prolapse in the community. Circulation 106, 1355–1361 (2002)
Freed, L. A. et al. Prevalence and clinical outcome of mitral-valve prolapse. N. Engl. J. Med. 341, 1–7 (1999)
Vaishnava, P., Fuster, V., Goldman, M. & Bonow, R. O. Surgery for asymptomatic degenerative aortic and mitral valve disease. Nat. Rev. Cardiol. 8, 173–177 (2011)
Waller, B. F., Maron, B. J., Del Negro, A. A., Gottdiener, J. S. & Roberts, W. C. Frequency and significance of M-mode echocardiographic evidence of mitral valve prolapse in clinically isolated pure mitral regurgitation: analysis of 65 patients having mitral valve replacement. Am. J. Cardiol. 53, 139–147 (1984)
Freed, L. A. et al. A locus for autosomal dominant mitral valve prolapse on chromosome 11p15.4. Am. J. Hum. Genet. 72, 1551–1559 (2003)
Levine, R. A. et al. Three-dimensional echocardiographic reconstruction of the mitral valve, with implications for the diagnosis of mitral valve prolapse. Circulation 80, 589–598 (1989)
Levine, R. A., Stathogiannis, E., Newell, J. B., Harrigan, P. & Weyman, A. E. Reconsideration of echocardiographic standards for mitral valve prolapse: lack of association between leaflet displacement isolated to the apical four chamber view and independent echocardiographic evidence of abnormality. J. Am. Coll. Cardiol. 11, 1010–1019 (1988)
Perloff, J. K. & Child, J. S. Clinical and epidemiologic issues in mitral valve prolapse: overview and perspective. Am. Heart J. 113, 1324–1332 (1987)
Clark, H. F. et al. Dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila . Genes Dev. 9, 1530–1542 (1995)
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nature Methods 7, 248–249 (2010)
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009)
Schwarz, J. M., Rodelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods 7, 575–576 (2010)
Visel, A., Thaller, C. & Eichele, G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556 (2004)
Guénette, S. et al. Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J. 25, 420–431 (2006)
Margolin, D. H. et al. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N. Engl. J. Med. 368, 1992–2003 (2013)
Golzio, C. et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485, 363–367 (2012)
Chaki, M. et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150, 533–548 (2012)
Norris, R. A. et al. Expression of the familial cardiac valvular dystrophy gene, filamin-A, during heart morphogenesis. Dev. Dyn. 239, 2118–2127 (2010)
Rabkin, E. et al. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation 104, 2525–2532 (2001)
Nesta, F. et al. Leaflet concavity: a rapid visual clue to the presence and mechanism of functional mitral regurgitation. J. Am. Soc. Echocardiogr. 16, 1301–1308 (2003)
Mao, Y. et al. Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development 138, 947–957 (2011)
Nesta, F. et al. New locus for autosomal dominant mitral valve prolapse on chromosome 13: clinical insights from genetic studies. Circulation 112, 2022–2030 (2005)
Cho, E. & Irvine, K. D. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development 131, 4489–4500 (2004)
Wessels, A. et al. Epicardially-derived fibroblasts and their contribution to the parietal leaflets of the atrioventricular valves in the murine heart. Dev. Biol. 366, 111–124 (2012)
Zuppiroli, A., Roman, M. J., O'Grady, M. & Devereux, R. B. A family study of anterior mitral leaflet thickness and mitral valve prolapse. Am. J. Cardiol. 82, 823–826 A10 (1998)
Kolibash, A. J., Jr et al. Evidence for progression from mild to severe mitral regurgitation in mitral valve prolapse. Am. J. Cardiol. 58, 762–767 (1986)
Li, K. & Stockwell, T. B. VariantClassifier: a hierarchical variant classifier for annotated genomes. BMC Res. Notes 3, 191 (2010)
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010)
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
Cooper, G. M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005)
The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Morrison, T. B., Weis, J. J. & Wittwer, C. T. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24, 954–958,–960, 962 (1998)
Thisse, B. et al. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519 (2004)
Kolpa, H. J. et al. miR-21 represses Pdcd4 during cardiac valvulogenesis. Development 140, 2172–2180 (2013)
Tsukasaki, Y. et al. Giant cadherins Fat and Dachsous self-bend to organize properly spaced intercellular junctions. Proc. Natl Acad. Sci. USA 111, 16011–16016 (2014)
Norris, R. A. et al. Periostin regulates atrioventricular valve maturation. Dev. Biol. 316, 200–213 (2008)
Sauls, K. et al. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovasc. Res. 96, 109–119 (2012)
Xu, F., Beyazoglu, T., Hefner, E., Gurkan, U. A. & Demirci, U. Automated and adaptable quantification of cellular alignment from microscopic images for tissue engineering applications. Tissue Eng. Part C Methods 17, 641–649 (2011)
Norris, R. A. et al. Identification and detection of the periostin gene in cardiac development. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 281A, 1227–1233 (2004)
Acknowledgements
This work was supported by the Fondation Leducq (Paris, France) Mitral Transatlantic Network of Excellence grant 07CVD04. MVP patient studies were supported by an Innovation in Clinical Research award of the Doris Duke Charitable Foundation, by an award of the Aetna Quality Care Research Fund, and by a gift from Rena M. Shulsky, New York, New York (S.A.S. and R.A.L.). Sequencing of the candidate region was performed at the Venter Institute through a grant from the National Heart Lung and Blood Institute Resequencing and Genotyping (RS&G) Service (S.A.S.). The work at MUSC was performed in a facility constructed with support from the National Institutes of Health, Grant Number C06 RR018823 from the Extramural Research Facilities Program of the National Center for Research Resources. Collection of the MVP France cohort was supported by the French Society of Cardiology. Other funding sources: National Heart Lung and Blood Institute: R01HL122906-01 (A.W.), R01-HL33756 (R.R.M.), COBRE 1P30 GM103342 (R.R.M., R.A.N., A.W.), 8P20 GM103444-07 (R.R.M. and R.A.N.), R01-HL109004 (D.J.M.), R01-HL127692 (D.J.M., S.A.S., R.A.N., R.A.L.); RO1-HL095696 (D.R.M.), VA Merit Review BX002327 (D.R.M.); National Institute of Mental Health R00-MH095867 (M.E.T.) The Hassenfeld Scholar Program (D.J.M.); The March of Dimes (M.E.T.); M.G.H. Scholars Program (S.A.S., M.E.T.); American Heart Association: 09GRNT2060075 (A.W.), 11SDG5270006 (R.A.N.), 2261354 (D.J.M.), 15GRNT25080052 (R.A.N.); National Science Foundation: EPS-0903795 (R.R.M.); NHLBI K24 HL67434, R01HL72265 and R01HL109506 and the Ellison Foundation, Boston, MA (R.A.L.), Howard Hughes Medical Institute (K.D.I.), and a gift from Michael Zak (D.J.M.). Thanks to T. Brown (MUSC) for his guidance on MRI studies, C. Hanscom (M.G.H.) for assistance with genomic libraries, and E. Lim (H.M.S.) for contributions in interpreting the exome mutation data. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Author information
Authors and Affiliations
Contributions
R.D., M.L., C.S., C.J., M.P., X.J., J.-J.S., D.T., S.O., X.E., F.C., and S.A.S. participated in genetic analysis, sequencing and mutation cloning. R.D., F.N.D., L.A.F., T.L.T., H.L.M., L.F.-F., J.S., C.T., R.A.L., and A.H. participated in patient collection and phenotyping using echocardiography. D.S.P., S.N.L. and D.J.M. performed zebrafish and cell culture experiments. M.E.T., M.R.S., N.B.N., C.D., H.B. and C.C. performed bioinformatics and statistical analysis. A.C., P.C., and P.B. established human patient cell cultures and histology on human tissues. A.d.V., K.W., K.D.I., Y.M., K.S., A.W., T.M., K.T., R.R.M. and R.A.N. performed mouse embryo and knockout experiments and cell alignment and migration assays. X.N., A.-M.B., D.R.M., H.K. performed mouse in vivo imaging. R.D., D.J.M., R.A.L., R.A.N. and S.A.S. wrote the manuscript. R.A.L., A.H., S.A.S. and J.J.S. coordinated the Leducq Mitral Network.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Measurement of endogenous and exogenous gene expression in D. rerio.
a, Corresponding representative embryos of each morpholino knockdown on the left, with close-up of heart on the right. b, To assess efficiency of morpholino knockdown, 20 embryos were collected 72 h after injection, mRNA was collected, and quantitative PCR was performed with three technical replicates. We demonstrate that morpholino (MO) knockdown of each indicated gene results in reduced mRNA expression, after normalization to beta-actin expression, compared to mock-injected controls (two-sided Student’s t-test). P values are noted on graphs. c, Western blotting of 20 pooled embryos injected with DCHS1 mRNA demonstrates the production of protein. Mutant mRNA refers to the compound mutant P197L/R2513H.
Extended Data Figure 2 Apbb1 is not expressed during cardiac morphogenesis.
Apbb1 RNA expression was analysed at E14.5 in sagittal sections using 2 separate antisense probes. Whereas strong cranial and neural expression is observed for Apbb1, no detectable cardiac expression or valve expression (arrow) is evident.
Extended Data Figure 3 Dachsous1b expression at the atrioventricular junction.
In situ hybridization reveals the presence of dchs1b in the atrioventricular canal (avc) at 54 hpf (a, b) and 72 hpf (c). The dchs1b expression is purple while a counterstain for cardiac tissue is brown (a). White arrows highlight the dchs1b signal in the atrioventricular canal.
Extended Data Figure 4 Dachsous1b knockdown alters atrioventricular ring markers.
In situ hybridization at 48 hpf and 72 hpf, as indicated, was performed for known atrioventricular ring markers. In contrast to WT (a) bmp4 expression is expanded into the ventricle at 48 hpf in dchs1 knockdown embryos at 48 hpf (b), spp1 and notch1b expression was largely unperturbed (c–f), and has2 expression was not detected at 48 hpf, and is faint at 72 hpf in dchs1 knockdown, compared to identically handled and stained controls (i–l).
Extended Data Figure 5 Histopathology mitral valves.
Human posterior leaflets of control, Barlow’s with MVP, and DCHS1 p.R2330C were isolated, fixed and stained with Movat’s pentachrome. Leaflet thickening, elongation and myxomatous degeneration is observed in the Barlow’s and DCHS1 p.R2330C leaflets compared to controls. Expansion of the proteoglycan layer (blue) and disruption of the normal stratification of matrix boundaries is observed in the Barlow’s and DCHS1 p.R2330C leaflets. Blue, proteoglycan; yellow, collagen; black, elastin; red, fibrin or cardiac muscle. Scale bars, 0.5 cm.
Extended Data Figure 6 Protein expression of uncoupled mutations.
In order to determine which family 1 DCHS1 mutation is leading to the observed decrease in protein expression, constructs were generated that harboured only the p.P197L or the p.R2513H variant. Mutant refers to the double mutant P197L/R2513H construct. Western blot analyses from transfected HEK293 cells, three independent biological replicates, demonstrate that the p.R2513H mutation causes a significant decrease in DCHS1 protein expression, similar to that of the construct with both variants (mutant), suggesting pathogenicity. Percent difference in protein levels is depicted. Normalization of data was accomplished by qPCR specific to the transfected constructs. P values from the Student’s t-test are indicated in graphs.
Extended Data Figure 7 Cardiac function is not altered in Dchs1+/− mice.
M-mode analyses were performed to determine whether cardiac structure and/or function were perturbed in the Dchs1+/− mice. No statistically significant differences were observed in either cardiac structure or calculated cardiac function (n = 6 for each genotype). IVS, interventricular septum; d, diastole; s, systole; LVID, left ventricular internal dimension; LVPW, left ventricular posterior wall; EF, ejection fraction; FS, fractional shortening; LV, left ventricle.
Extended Data Figure 8 Dchs1 expression during cardiac development.
Top, RNA expression of Dchs1 was analysed during embryonic gestation (E11.5, E13.5, and E15.5) by section in situ hybridization. At E11.5 Dchs1 RNA (blue staining) expression is observed in the endocardium and mesenchyme of the superior and inferior cushions (sAVC and iAVC, respectively). A gradient pattern of expression is observed at this time point with more intense expression near the endocardium. At E13.5 and E15.5, a similar pattern is observed in the forming anterior and posterior mitral leaflets (AL and PL, respectively). Bottom, Dchs1 protein expression (red) is observed throughout cardiac development in the endothelial cells and interstitial cells of the developing valves. Dchs1 shows asymmetric expression in the valvular interstitial cell bodies around E15.5 (arrowheads). Dchs1 protein is also observed in the epicardium and atrioventricular sulcus (arrows). (Red Dchs1; green MF20; blue Hoescht).
Extended Data Figure 9 Dchs1 deficiency causes altered valvular interstitial cell patterning in vivo.
a, IHC for eGFP of postnatal day 0 (P0) lineage traced Wt1-Cre/Rosa-eGFP/Dchs1+/+ neonatal mice show epicardial-derived cells (EPDCs) migrating into the posterior leaflet as a sheet of cells directly under the endothelium of the atrialis. This normal patterning is perturbed in the Wt1-Cre/Rose-eGFP/Dchs1+/− mice. 3D reconstructions were used to examine all EPDCs in the posterior leaflet of both genotypes to obtain a complete fate map of these cells. b, c, Total volume of the leaflet is unchanged at this time point. However, the total volume of EPDCs as well as total EPDC cell number is significantly increased. There is a significant decrease in the number of non-EPDCs in the posterior leaflet with no overall change in total cell number. These data demonstrate that a minimum threshold of Dchs1 expression is required for normal migration of EPDCs into the posterior leaflet, normal patterning of this cell population, and cross-talk between EPDC and non-EPDC cell types in the valve. **P < 0.01. d, Isolated anterior mitral leaflet from fetal (E17.5) Dchs1+/+ , Dchs1+/− , and Dchs1−/− mice were used to quantify cellular alignment of valvular interstitial cells. Vector maps were generated from histological (haematoxylin and eosin) stains to show orientation and alignment of cells in relationship to each other. Boxes in each vector map panel are represented as zoomed images of regions within each of the valves to show cell orientation. e, Cell alignment and polarity were quantified as the number of cells that deviate >10 degrees from the proximal-distal (P–D) axis of the leaflet. 90% of the cells in Dchs1+/+ show proper alignment with each other and along this P–D axis. Haploinsufficiency (Dchs1+/− ) results in a 50% reduction in cell alignment, which is further reduced in Dchs1−/− (*P values < 0.01).
Extended Data Figure 10 Mice and MVP patients with Dchs1 deficiency exhibit migratory defects in vitro.
a, Posterior leaflets of P0 neonatal Dchs1+/+ and Dchs1+/− mice were explanted and interstitial cells were allowed to migrate out for 24 h. Dchs1+/− mice exhibit increased migration (black lines drawn from explants) coincident with loss of cell–cell contacts and N-cadherin expression at focal adhesions. Whereas N-cadherin expression (red) is found at the membrane at points of cell–cell contract in Dchs1+/+ valvular interstitial cells (arrows), this membrane expression is lost in the Dchs1+/− cells and is prominently expressed in the cytoplasm (arrows). Nuclei, blue. b, Migration assays using control and MVP patient (p.R2330C) valvular interstitial cells exhibit a similar affect as observed in the mouse cells whereby the p.R2330C cells exhibit an increase in migration. P values are indicated in graphs.
Supplementary information
Parasternal long-axis view of family 1 proband
This video shows posterior leaflet prolapse and dilated left-heart chambers. (MOV 1102 kb)
Zoomed view of thickened, prolapsing leaflet in family 1 proband.
This video shows thickened, prolapsing leaflet in family 1 proband. (MOV 684 kb)
Doppler color flow mapping of family proband 1
This video shows severe MR, increasing with prolapse throughout systole. (MOV 808 kb)
High-speed video of wild type (control injected) D. rerio heart at 72 hours post-fertilization (hpf).
At this stage the heart has looped and blood flow is unidirectional from the atrium (upper right) to the ventricle, and a constriction has formed at the junction between the atrium and ventricle. No regurgitation is evident. (AVI 6980 kb)
High-speed video of dchs1b morphant D. rerio heart at 72 hpf.
Hearts of dchs1b morphants fail to loop properly and there is regurgitation of blood from the ventricle (lower right) into the atrium (upper left). Concomitant with this phenotype, there is reduced constriction of the AV canal. (AVI 6833 kb)
Parasternal long axis view of adult (9-month old) Dchs1+/+ heart
This video shows normal mitral valve opening and closing. (MP4 16824 kb)
Parasternal long-axis view of adult (9-month old) Dchs1+/- heart
This video shows anterior and posterior leaflet thickening and posterior leaflet prolapse. Prolapse is most easily observed at frames: 149, 188, and 300 (MP4 16671 kb)
This video comprises 2 clips, which show 3D reconstructions of Dchs1+/+ 9, and Dchs1+/- 9-month old posterior leaflet
Each of the clips shown were obtained from the micro-MRI slices. (MP4 16062 kb)
AMIRA 3D reconstruction of Dchs1+/+, Dchs1+/-, and Dchs1-/- E17.5 mitral leaflets.
Respective 3D reconstructions are shown sequentially during the movie. Green=Anterior Leaflet, Blue =Posterior Leaflet. (MP4 23413 kb)
AMIRA 3D reconstruction of EPDC lineage trace in Dchs1+/+ and Dchs1+/- PO posterior mitral leaflets.
Respective 3D reconstructions are shown sequentially during the movie. Green=EPDC, Blue =non-EPDCs (MP4 16431 kb)
Rights and permissions
About this article
Cite this article
Durst, R., Sauls, K., Peal, D. et al. Mutations in DCHS1 cause mitral valve prolapse. Nature 525, 109–113 (2015). https://doi.org/10.1038/nature14670
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14670
This article is cited by
-
Valvular heart disease and cardiomyopathy: reappraisal of their interplay
Nature Reviews Cardiology (2024)
-
Degenerative mitral regurgitation
Nature Reviews Disease Primers (2023)
-
A change of heart: new roles for cilia in cardiac development and disease
Nature Reviews Cardiology (2022)
-
Single cell transcriptomics and TCR reconstruction reveal CD4 T cell response to MHC-II-restricted APOB epitope in human cardiovascular disease
Nature Cardiovascular Research (2022)
-
Phenotypes of Cardiovascular Diseases: Current Status and Future Perspectives
Phenomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.