Abstract
Bacteria share their ecological niches with other microbes. The bacterial type VI secretion system is one of the key players in microbial competition, as well as being an important virulence determinant during bacterial infections. It assembles a nano-crossbow-like structure in the cytoplasm of the attacker cell that propels an arrow made of a haemolysin co-regulated protein (Hcp) tube and a valine–glycine repeat protein G (VgrG) spike and punctures the prey’s cell wall. The nano-crossbow is stably anchored to the cell envelope of the attacker by a membrane core complex. Here we show that this complex is assembled by the sequential addition of three type VI subunits (Tss)—TssJ, TssM and TssL—and present a structure of the fully assembled complex at 11.6 Å resolution, determined by negative-stain electron microscopy. With overall C5 symmetry, this 1.7-megadalton complex comprises a large base in the cytoplasm. It extends in the periplasm via ten arches to form a double-ring structure containing the carboxy-terminal domain of TssM (TssMct) and TssJ that is anchored in the outer membrane. The crystal structure of the TssMct–TssJ complex coupled to whole-cell accessibility studies suggest that large conformational changes induce transient pore formation in the outer membrane, allowing passage of the attacking Hcp tube/VgrG spike.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The EM structure of the TssJLM complex has been deposited in the Electron Microscopy Data Bank (EMDB) under accession number emd-2927. The crystal structures of the TssM32Ct–nb25 complex, and of the TssM26Ct fragment and TssM26Ct–TssJ complexes, have been deposited in the Protein Data Bank under accession numbers 4Y7M, 4Y7L and 4Y7O, respectively.
References
West, S. A., Griffin, A. S. & Gardner, A. Evolutionary explanations for cooperation. Curr. Biol. 17, 661–672 (2007)
Blango, M. G. & Mulvey, M. A. Bacterial landlines: contact-dependent signaling in bacterial populations. Curr. Opin. Microbiol. 12, 177–181 (2009)
Russell, A. B., Peterson, S. B. & Mougous, J. D. Type VI secretion system effectors: poisons with a purpose. Nature Rev. Microbiol. 12, 137–148 (2014)
Silverman, J. M., Brunet, Y. R., Cascales, E. & Mougous, J. D. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472 (2012)
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D. & Mekalanos, J. J. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl Acad. Sci. USA 104, 15508–15513 (2007)
Russell, A. B. et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 (2011)
Russell, A. B. et al. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512 (2013)
Ma, L. S., Hachani, A., Lin, J. S., Filloux, A. & Lai, E. M. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104 (2014)
Durand, E., Cambillau, C., Cascales, E., Journet, L. & Vgr, G. Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507 (2014)
Borgeaud, S., Metzger, L. C., Scrignari, T. & Blokesch, M. Bacterial evolution. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015)
Cascales, E. The type VI secretion toolkit. EMBO Rep. 9, 735–741 (2008)
Bönemann, G., Pietrosiuk, A., Diemand, A., Zentgraf, H. & Mogk, A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 (2009)
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J. & Mekalanos, J. J. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012)
Kudryashev, M. et al. Structure of the type VI secretion system contractile sheath. Cell 160, 952–962 (2015)
Coulthurst, S. J. The type VI secretion system – a widespread and versatile cell targeting system. Res. Microbiol. 164, 640–654 (2013)
Ho, B. T., Dong, T. G. & Mekalanos, J. J. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21 (2014)
Zoued, A. et al. Architecture and assembly of the type VI secretion system. Biochim. Biophys. Acta 1843, 1664–1673 (2014)
Bönemann, G., Pietrosiuk, A. & Mogk, A. Tubules and donuts: a type VI secretion story. Mol. Microbiol. 76, 815–821 (2010)
Brunet, Y. R., Hénin, J., Celia, H. & Cascales, E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 15, 315–321 (2014)
Shneider, M. M. et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353 (2013)
Brunet, Y. R., Espinosa, L., Harchouni, S., Mignot, T. & Cascales, E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 3, 36–41 (2013)
Aschtgen, M. S., Gavioli, M., Dessen, A., Lloubès, R. & Cascales, E. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol. Microbiol. 75, 886–899 (2010)
Aschtgen, M. S., Bernard, C. S., de Bentzmann, S., Lloubès, R. & Cascales, E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531 (2008)
Ma, L. S., Lin, J. S. & Lai, E. M. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329 (2009)
Felisberto-Rodrigues, C. et al. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7, e1002386 (2011)
Ma, L. S., Narberhaus, F. & Lai, E. M. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287, 15610–15621 (2012)
Aschtgen, M. S., Zoued, A., Lloubès, R., Journet, L. & Cascales, E. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 type VI secretion system, is inserted by YidC. MicrobiologyOpen 1, 71–82 (2012)
Durand, E. et al. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J. Biol. Chem. 287, 14157–14168 (2012)
Chang, J. H. & Kim, Y. G. Crystal structure of the bacterial type VI secretion system component TssL from Vibrio cholerae. J. Microbiol. 53, 32–37 (2015)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Nguyen, V. S. et al. Production, crystallization and X-ray diffraction analysis of a complex between a fragment of the TssM T6SS protein and a camelid antibody. Acta Crystallogr F. 71, 266–271 (2015)
Nguyen, V. S. et al. Inhibition of type VI secretion by an anti-TssM llama nanobody. PLoS ONE 10, e0122187 (2015)
Diepold, A. et al. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29, 1928–1940 (2010)
Judd, P. K., Kumar, R. B. & Das, A. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl Acad. Sci. USA 102, 11498–11503 (2005)
Hardie, K. R., Lory, S. & Pugsley, A. P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15, 978–988 (1996)
Drake, S. L., Sandstedt, S. A. & Koomey, M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol. 23, 657–668 (1997)
Burghout, P. et al. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J. Bacteriol. 186, 5366–5375 (2004)
Crago, A. M. & Koronakis, V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30, 47–56 (1998)
Daefler, S. & Russel, M. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28, 1367–1380 (1998)
Brunet, Y. R., Bernard, C. S., Gavioli, M., Lloubès, R. & Cascales, E. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7, e1002205 (2011)
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000)
Chaveroche, M. K., Ghigo, J. M. & d’Enfert, C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28, e97 (2000)
van den Ent, F. & Löwe, J. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67–74 (2006)
Gueguen, E. & Cascales, E. Promoter swapping unveils the role of the Citrobacter rodentium CTS1 type VI secretion system in interbacterial competition. Appl. Environ. Microbiol. 79, 32–38 (2013)
Zaslaver, A. et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nature Methods 3, 623–628 (2006)
Zoued, A. et al. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J. Biol. Chem. 288, 27031–27041 (2013)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Scheres, S. H. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189, 114–122 (2015)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nature Methods 11, 63–65 (2014)
Pettersen, E. F. et al. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. & Svergun, D. I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003)
Konarev, P. V., Petoukhov, M. V., Volkov, V. V. & Svergun, D. I. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 (2006)
Guinier, A. La diffraction des rayons X aux très petits angles; application à l’étude de phénomènes ultramicroscopiques. Ann. Phys. (Paris) 12, 161–237 (1939)
Svergun, D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992)
Franke, D. & Svergun, D. I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009)
Volkov, V. V. & Svergun, D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003)
Kozin, M. B. & Svergun, D. I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 (2001)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D 66, 22–25 (2010)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Winn, M. D., Murshudov, G. N. & Papiz, M. Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
The PyMOL Molecular Graphics System. v.1.5.0.4 (Schrödinger, LLC, 2014)
Chapman, M. S., Trzynka, A. & Chapman, B. K. Atomic modeling of cryo-electron microscopy reconstructions – joint refinement of model and imaging parameters. J. Struct. Biol. 182, 10–21 (2013)
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Bogdanov, M., Zhang, W., Xie, J. & Dowhan, W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAMTM): application to lipid-specific membrane protein topogenesis. Methods 36, 148–171 (2005)
Goemaere, E. L., Devert, A., Lloubès, R. & Cascales, E. Movements of the TolR C-terminal domain depend on TolQR ionizable key residues and regulate activity of the Tol complex. J. Biol. Chem. 282, 17749–17757 (2007)
Du, D. et al. Structure of the AcrAB–TolC multidrug efflux pump. Nature 509, 512–515 (2014)
Hodgkinson, J. L. et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nature Struct. Mol. Biol. 16, 477–485 (2009)
Low, H. H. et al. Structure of a type IV secretion system. Nature 508, 550–553 (2014)
Acknowledgements
This work was funded by Agence Nationale de la Recherche (ANR) grants ANR-10-JCJC-1303-03 to E.C., Bip:Bip to R.F., ANR-14-CE14-0006-02 to C.C. and E.C., and supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05-01. E.D. was supported by a post-doctoral fellowship from the Fondation pour la Recherche Médicale (SPF20101221116) and ANR grants Bip:Bip and ANR-10-JCJC-1303-03. V.S.N. was supported by a PhD grant from the French Embassy in Vietnam (792803C). A.Z., L.L. and M.S.A. were recipients of doctoral fellowships from the French Ministère de la Recherche. A.Z. received a Fondation pour la Recherche Médicale fellowship (FDT20140931060). We thank O. Francetic for providing anti-DglA and anti-OmpF antibodies. We thank E. Marza, P. Violinova Krasteva and H. Remaut for comments on the manuscript, and T. Mignot, M. Guzzo and L. Espinosa for advice about the fluorescence microscopy experiments and the statistical analyses. We also thank the members of the R.F. and E.C. research groups for discussions and suggestions, and R. Lloubès, J. Sturgis and A. Galinier for encouragement. We thank the ERSF and Soleil Synchrotron radiation facilities for beamline allocation.
Author information
Authors and Affiliations
Contributions
E.D., A.Z., C.C., E.C. and R.F. designed the experiments. A.Z. constructed the EAEC mutant and fluorescent strains and performed the fluorescence microscopy experiments and statistical analyses. L.L. and M.S.A. constructed the TssM cysteine derivatives and performed the accessibility experiments. E.D. assisted by An.D. purified the TssJLM complex and performed its biochemical characterization. E.D. and G.P.A. collected the EM data. E.D. and R.F. obtained the 3D reconstruction of the TssJLM complex. V.S.N., S.S., A.R. and C.C. purified, crystallized and solved the X-ray structures. Al.D. generated the nanobody. B.B. obtained the energy-minimized models of the closed and open states of the TssM26Ct–TssJ complex.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Functional and dynamic properties of fluorescently labelled Tss proteins.
a, GFP–TssM and GFP–TssL fusion proteins are functional. Top: Hcp release assay. Hcp release was assessed by separating whole cells (C) and supernatant (S) fractions from the indicated strains. A total of 1 × 109 cells and the TCA-precipitated material from the supernatant of 2 × 109 cells were analysed by western blot using anti-Flag monoclonal antibody (lower panel) and anti-TolB polyclonal antibodies as a lysis control (upper panel). The molecular mass markers (in kilodaltons) are indicated on the left. Bottom: anti-bacterial assay. The anti-bacterial activity was assessed by mixing kanamycin-resistant prey E. coli K-12 cells with the indicated attacker cells for 16 h at 37 °C in SIM. The number of recovered E. coli prey cells is indicated in the graph (as log of colony-forming units (c.f.u.)). The circles indicate values from three independent assays, and the average is indicated by the bar. b, TssJ–sfGFP and TssJ–mCh fusion proteins are non-functional. Hcp release was assessed by separating whole cells (C) and supernatant (S) fractions from the indicated strains. A total of 1 × 109 cells and the TCA-precipitated material from the supernatant of 2 × 109 cells were analysed by western blot using anti-Flag monoclonal antibody (lower panel) and anti-TolB polyclonal antibodies as a lysis control (upper panel). The molecular mass markers (in kilodaltons) are indicated on the left. c, sfGFP–TssM and sfGFP–TssL cluster in foci. Large fields of fluorescence microscopy recordings showing localization of the sfGFP–TssL (left) and sfGFP–TssM (right) fusion proteins. The positions of selected foci are indicated by arrowheads. Scale bars, 5 μm. d, sfGFP–TssM and sfGFP–TssL foci are stable and static. Mean square displacement (in arbitrary units (a.u.)) of sfGFP–TssM (blue line) and sfGFP–TssL (red line) clusters were measured by sub-pixel tracking of fluorescent foci and plotted over time (in minutes). e, The TssBC sheath tubular structures assemble on TssJLM membrane complexes. Statistical analyses reporting the average number of sheath per cell compared with the number of membrane complexes per cell, highlighting the observation that the number of membrane complexes is at least equal to the number of sheathes. Lower and upper boundaries of the boxes correspond to the 25% and 75% percentiles respectively. Black bold horizontal bar, median values for each strain; whiskers, 10% and 90% percentiles; n indicates the number of cells studied per strain. f, Long-term fluorescence microscopy recordings. Time-lapse fluorescence microscopy recordings showing localization and dynamics of the sfGFP–TssL and TssB–mCherry fusion proteins. Individual images were taken every 15 min. Assembly/contraction of the sheath and TssL localization events are schematized in the lowest panel. Scale bars, 1 μm. g, Statistical analysis of sfGFP–TssM and sfGFP–TssL localization in various tss backgrounds. Shown are box-and-whisker plots of the measured number of sfGFP–TssM and sfGFP–TssL foci per cell for each indicated strain with the lower and upper boundaries of the boxes corresponding to the 25% and 75% percentiles respectively (horizontal bar, the median values for each strain; whiskers, the 10% and 90% percentiles); n indicates the number of cells studied per strain.
Extended Data Figure 2 Expression and purification of the T6SS membrane core complex.
a–e, T6SS operon genomic organization and constructs used for in vitro analyses. a, Schematic representation of the T6SS sci-1 gene cluster from entero aggregative E. coli. The numbers on top refer to the gene locus tag (EC042_XXXX). Genes encoding core components (identified by their names on bottom, for example, ‘B’ refers to the tssB gene) are coloured grey. Genes of unknown function are coloured white. The three genes used to reconstitute the core membrane complex are coloured orange (tssJ), blue (tssL) and green (tssM). b, Schematic representation of the engineered constructs: the tssJ, tssL and tssM genes were amplified with an additional Shine Dalgarno (SD) sequence and 3′ StrepII, 5′ Flag and 5′ 6×His tags respectively. These three fragments were cloned into the pRSF-Duet vector (c). This construct allows the production of the C-terminally StrepII-tagged TssJ outer membrane (OM) lipoprotein and N-terminally Flag-tagged TssL and 6×His-tagged TssM inner-membrane (IM) proteins (d, e). The proteins are schematized and their boundaries and principal characteristics (TM, transmembrane segments; SP, signal peptide; CYS, acylated cysteine) are indicated (d) and their topologies are shown (e). The additional TssM constructs (TssMp, TssM32Ct and TssM26Ct) used for SAXS or X-ray analyses are shown at the bottom. f–h, Purification and biochemical characterization of the T6SS membrane core complex. f, Analytical size-exclusion chromatography analysis of the purified TssJLM complex (continuous line) on a Superose 6 column, calibrated with 75-, 158-, 440- and 660-kDa molecular mass markers (dotted lines). The molecular mass of each marker (in kilodaltons) is indicated on the top of the corresponding peak. An arrow indicates the position of the peak fraction corresponding to the TssJLM complex. g, SDS–PAGE of the purified TssJLM complex analysed by Coomassie staining (CB) or immunoblotting using anti-His (α-His), -Flag (α-Flag) and -StrepII (α-STREP) antibodies. h, Left: cysteine labelling of the purified TssJLM complex in reducing and denaturing conditions as described in Methods. The total number of cysteine residues was nine for TssM, five for TssL and none for TssJ (the N-terminal cysteine is acylated). Right: the relative amount of TssL compared with TssM (densitometry relative to the number of free cysteine residues, fixed at 1 for TssM).
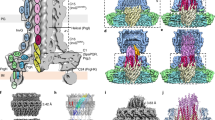
Extended Data Figure 3 Architecture of the T6SS membrane core complex.
a, Negative-stain EM of the EAEC TssJLM complex. Representative micrograph of the data set used for image processing. Isolated TssJLM complexes were clearly visible (white circles). b, Plot of the rotational autocorrelation function for a representative class average of an end-view. c, FSC curve of the TssJLM reconstruction. The ‘gold standard’ FSC curve was calculated in Relion using the masked reconstruction of the TssJLM complex. The resolution at 0.143 correlation was 11.56 Å. d, Top: side and corresponding cut-away views of the 3D reconstruction for the whole TssJLM complex. Bottom: local resolution as calculated by Resmap. The TssJLM volume (left reconstruction, side view; right reconstruction, cut-away view) is coloured according to the local resolution from high resolution (∼12 Å) in blue to low resolution (>30 Å) in red. e, FSC curve of the TssJLM base. The ‘gold standard’ FSC curve was calculated in Relion using the unmasked reconstruction of the TssJLM base. The resolution at 0.143 correlation was 16.6 Å. f, Top, side and bottom views of the 3D reconstruction after specific refinement of the base.
Extended Data Figure 4 Structural analysis and segmentation.
a, Segmentation of the TssJLM reconstruction. Left: above the base, ten equivalent densities could be defined by segmentation. They are arranged in two concentric rings. The internal ring is represented in green in the top panel and the external ring is represented in blue in the bottom panel. Right: cut-out views of the complex showing the arrangement of the two concentric rings at different levels (grey lines) along the periplasmic portion of the TssJLM complex. The cut-out views are seen from the bottom of the complex. b, Requirement of TssJ lipidation for complex assembly and insertion into the outer membrane. Left: membrane fractionation by differential solubilization followed by immunoblot analysis. Total membrane extracts from cells producing the wild-type TssJLM complex or the TssJLM complex with an unacylated variant of TssJ (Cys1-to-Ser substitution, CS) were solubilized by lauroyl sarcosine to separate inner membranes and outer membranes. HisTssM, FlagTssL and TssJStrep (indicated on the right) were revealed by anti-His, anti-Flag and anti-StrepII antibodies respectively. Controls included immuno-detection of the inner membrane DglA diacylglycrol lipase and the outer membrane OmpF porin. Wild-type TssJLM complex co-fractionates with both the inner and outer membrane fractions whereas the Cys1-to-Ser substitution mutated complex co-fractionates only with the inner-membrane fraction. Top right: negative-stain EM of the mutated TssJCSLM complex. Representative micrograph of the data set used for image processing. Isolated TssJCSLM complexes were clearly visible (white circles). Bottom right: gallery of representative class averages generated after reference-free 2D classification in Relion. End to side views are shown from top left to bottom right. c, Orientation of the TssJLM complex in the cell envelope. Left: schematic representation of the TssJ (J, orange), TssL (L, blue) and TssM (M, green) proteins. Their localization, main characteristics (lipidation or transmembrane segments shown in black) and the location of the 6×His and StrepII tags (red balls) are indicated. The strepII and 6×His tags were introduced at the C terminus and N terminus of TssJ and TssM respectively. Middle: immune and Nanogold labelling coupled to EM. Anti-StrepII or Nanogold-NTA were incubated with the TssJLM complex and visualized by negative-stain EM. A gallery of representative views is presented (top row, StrepII labelling; bottom row, Ni-NTA labelling). StrepII antibodies (a schematic diagram with StrepII antibodies depicted as blue circles is shown on top) and Nanogold-NTA are highlighted in red circles. Right: the positions of the StrepII antibody (targeting TssJ C terminus) and of the Ni-NTA gold particle (targeting TssM N terminus) are indicated on the TssJLM reconstruction. d, SAXS data and low-resolution structure of the TssMp–TssJ complex. Top left: experimental scattering data (green crosses) and the fitting curve (continuous red line) calculated from an ab initio model of the TssMp–TssJ complex. Top right: Guinier plot (dots) with the linear fit (continuous line). Bottom left: distance distribution function of the TssMp–TssJ complex. Bottom right: SAXS envelope (grey surface) of the ‘best representative’ model of the TssMp–TssJ complex. Each view is rotated by 90° around the y-axis. e, Location of the TssMp–TssJ complex SAXS envelope in the 3D reconstruction of TssJLM complex. Left: the volume of the TssMp–TssJ complex determined by SAXS was docked into the EM 3D reconstruction of the TssJLM complex (top). Two optimal docking positions were found, both with 82% correlation with the EM map (coloured magenta and pink). The corresponding volumes in the EM map were extracted (bottom). They correspond to the same volume displayed in Extended Data Fig. 4a. Right: direct comparison of the SAXS (magenta) and EM (blue and green) volumes corresponding to the TssMp–TssJ complex. The volumes are equivalent in size and shape. f, TssL cytoplasmic domain docking into the TssJLM complex base. Fitting of the TssL cytoplasmic domain (TssLcyto)28 dimer in green ribbons in the hooks found in the base. Top and bottom: side and bottom views, respectively.
Extended Data Figure 5 Crystal structure of the TssJ–TssM C-terminal domain complex.
a, Amino-acid sequence of TssM. The different domains as well as the fragments used in this study are indicated (yellow, transmembrane helix; grey, cytoplasmic domain; green, blue and purple, periplasmic domain; blue and purple, C-terminal domain corresponding to the TssM32Ct fragment; purple, C-terminal domain corresponding to the TssM26Ct fragment). b, Crystal structure of the TssM32Ct–nb25 complex. The two proteins are represented as rainbow-coloured ribbons. The complementary determining regions (CDRs 1–3, coloured blue, green and red, respectively) of nb25 are indicated. The inset highlights the TssM32Ct–nb25 interface: the TssM32Ct surface is coloured beige whereas nb25 is represented as rainbow-coloured ribbons; the side chains of the amino acids in contact with TssM32Ct are indicated. The nb25 nanobody binds the TssM C-terminal domain, and covers a surface area of 580 Å2 by inserting its protruding CDR3 between TssM32Ct loops L5–6 and L9–10. The contacts between the two proteins are listed in Extended Data Table 2a. c, Crystal structure of the TssM26Ct–TssJ complex. Left: the two proteins are represented as ribbons and coloured in rainbow mode. Middle: same view rotated by 90°. The TssJ loop 1–2, previously shown to contact TssM25, is indicated. Right: TssM26Ct–TssJ interface. Top panel: the TssM26Ct surface is coloured violet, whereas TssJ is represented as rainbow-coloured ribbons. The TssJ side-chains of the amino acids in contact with TssM are indicated. The loops are numbered according to the flanking β-strands. Bottom panel: the TssJ surface is coloured beige whereas TssM26Ct is represented as rainbow-coloured ribbons. The TssM side-chains of the amino acids in contact with TssJ are indicated. The contacts between the two proteins are listed in Extended Data Table 2b. d, Comparison of the binding sites of nb25 and TssJ on TssM. Left: the structure of the TssM26Ct–TssJ complex (rainbow coloured) has been superimposed to the structure of the TssM32Ct–nb25 complex (only nb25 is shown in grey for clarity). Right: the same partners as in the left panel in surface representation. TssM26Ct (violet), TssJ (green) and nb25 (pink). e, Insertion of the TssJ lipid anchor in the outer membrane. Left: TssJ structure25 with the N-terminal 24 residues (absent in the crystal structure). This N-terminal extension (in magenta), predicted to be disordered, was modelled in Chimera using Modeller. The first cysteine residue is acylated to allow anchorage to the inner leaflet of the outer membrane (orange rectangle). Right: docking of the TssM26Ct–TssJ complex in the EM 3D reconstruction of the TssJLM complex (only the uppermost (tip) part of the TssJLM complex is shown). Left panel: two TssM26Ct–TssJ were docked into the inner and outer pillars of the tip complex. Right panel: docking in each pillar of the TssJLM tip complex (C5 symmetry). f, Hydrophobicity of the TssM26Ct–TssJ complex. Surface representation of the TssM26Ct–TssJ decamer (left, top view; right, side view). The hydrophobicity of the surface residues is displayed (blue to red scale from most hydrophilic to most hydrophobic). No obvious hydrophobic patch is visible at the surface of the complex.
Extended Data Figure 6 Cell surface accessibility of TssM C-terminal domain.
a, Functionality of the TssM cysteine variants. Hcp release was assessed by separating whole cells (C) and supernatant (S) fractions from the wild-type (WT) 17-2 strain and its ΔtssM derivative producing a wild-type allele of TssM or TssM cysteine substitution derivatives (as indicated). A total of 1 × 109 cells and the TCA-precipitated material from the supernatant of 2 × 109 cells were analysed by western blot using anti-HA monoclonal antibody (lower panel) and anti-TolB polyclonal antibodies as a lysis control (upper panel). The molecular mass markers (in kilodaltons) are indicated on the left. b, Cysteine substitution labelling. Accessibility to cysteine residues positioned in TssM domain 4 loops was assessed by treating isolated membranes (M) or whole cells (WC) of the indicated strain (WT, wild-type 17-2; ΔtssBC; ΔtssJ) producing the indicated TssM cysteine derivative (in red letters) with the cysteine-reactive, membrane-impermeant BSA-maleimide (BSA-mal). Samples corresponding to a total of 5 × 109 cells were analysed by western blot using anti-Flag monoclonal antibody. The position of the TssM protein (∼125 kDa) is indicated as well as that of a retarded band corresponding to BSA-maleimide-coupled TssM (∼190 kDa; asterisk). The molecular mass markers (in kilodaltons) are indicated. c, Close-up of the TssM26Ct–TssJ interface. TssM26Ct is represented in blue ribbons. TssJ is represented in orange ribbons and orange transparent surface. TssM residues accessible from the cell exterior when the T6SS is functional are indicated by yellow spheres whereas unaccessible residues are shown by grey spheres. The accessible residues 989 and 1005 are buried at the interface between TssM and TssJ, suggesting that this interface is probably disrupted during T6SS assembly and/or function. Left and right panels are orthogonal views of the same molecule.
Extended Data Figure 7 Comparison with other bacterial secretion systems and model for channel opening.
a, Comparison between the T6SS TssJLM membrane core complex structure and other bacterial secretion systems. From left to right, the E. coli AcrAB-TolC multi-drug efflux pump (EMDB accession number emd-5915)73, the EAEC T6SS membrane core complex (this study, EMDB accession number emd-2927), the Shigella T3SS transmembrane complex (EMDB accession number emd-1617)74 and the E. coli R388 T4SS complex (EMDB accession number emd-2567)75. The positions on the inner membrane (IM) and outer membrane (OM) are indicated (C, cytoplasm; P, periplasm). Scale bar, 10 nm. b, Docking of the Hcp tube/VgrG spike into the TssJLM 3D reconstruction. Left: before sheath contraction. The Hcp tube/VgrG spike (VgrG in yellow and Hcp in green; surface representation) was manually docked in the 3D reconstruction of TssJLM complex (grey surface). The diameter of the channel defined by the closed tip complex is not large enough to allow the passage of the tube/spike, suggesting that large conformational changes probably occur. The cavity at the tip of VgrG could be filled by VgrG-bound PAAR modules or toxin effectors20. Right: during sheath contraction. The diameter of the C10-symmetrized TssM26Ct model (represented as ribbons) is compatible with the passage of the Hcp tube/VgrG spike (same colours as in the left panel). c, Closed and open forms of the TssM26Ct oligomer. Crystal structure of TssM26Ct represented as ribbons and transparent surface. The TssM26Ct α- and β-domains are coloured cyan and blue, respectively. The C-terminal α5-helix and the extended stretch are coloured pink. Cysteines with extracellular accessibility when the T6SS is active are coloured yellow, while the unlabelled ones are coloured red. Left: docking of the TssM26Ct–TssJ crystal structure in the EM 3D reconstruction of the TssJLM tip complex. Top and bottom panels, side and top views, respectively. Right: model of a C10-symmetrized oligomer of the TssM26Ct domain. Top and bottom panels, side and top views, respectively.
Supplementary information
Supplementary Tables
This file contains Supplementary Table 1. (PDF 222 kb)
Rights and permissions
About this article
Cite this article
Durand, E., Nguyen, V., Zoued, A. et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523, 555–560 (2015). https://doi.org/10.1038/nature14667
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14667
This article is cited by
-
Cytoplasmic contractile injection systems mediate cell death in Streptomyces
Nature Microbiology (2023)
-
Comparative genomics revealed that Vibrio furnissii and Vibrio fluvialis have mutations in genes related to T6SS1 and T6SS2
Archives of Microbiology (2023)
-
The P. aeruginosa effector Tse5 forms membrane pores disrupting the membrane potential of intoxicated bacteria
Communications Biology (2022)
-
Abiotic factors modulate interspecies competition mediated by the type VI secretion system effectors in Vibrio cholerae
The ISME Journal (2022)
-
Type VI secretion system mutations reduced competitive fitness of classical Vibrio cholerae biotype
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.