Abstract
The plant hormone jasmonate plays crucial roles in regulating plant responses to herbivorous insects and microbial pathogens and is an important regulator of plant growth and development1,2,3,4,5,6,7. Key mediators of jasmonate signalling include MYC transcription factors, which are repressed by jasmonate ZIM-domain (JAZ) transcriptional repressors in the resting state. In the presence of active jasmonate, JAZ proteins function as jasmonate co-receptors by forming a hormone-dependent complex with COI1, the F-box subunit of an SCF-type ubiquitin E3 ligase8,9,10,11. The hormone-dependent formation of the COI1–JAZ co-receptor complex leads to ubiquitination and proteasome-dependent degradation of JAZ repressors and release of MYC proteins from transcriptional repression3,10,12. The mechanism by which JAZ proteins repress MYC transcription factors and how JAZ proteins switch between the repressor function in the absence of hormone and the co-receptor function in the presence of hormone remain enigmatic. Here we show that Arabidopsis MYC3 undergoes pronounced conformational changes when bound to the conserved Jas motif of the JAZ9 repressor. The Jas motif, previously shown to bind to hormone as a partly unwound helix, forms a complete α-helix that displaces the amino (N)-terminal helix of MYC3 and becomes an integral part of the MYC N-terminal fold. In this position, the Jas helix competitively inhibits MYC3 interaction with the MED25 subunit of the transcriptional Mediator complex. Our structural and functional studies elucidate a dynamic molecular switch mechanism that governs the repression and activation of a major plant hormone pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
09 September 2015
A formatting issue in Extended Data Fig. 4 was corrected.
References
Browse, J. Jasmonate: preventing the maize tassel from getting in touch with his feminine side. Sci. Signal. 2, pe9 (2009)
Wager, A. & Browse, J. Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 3, 41 (2012)
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007)
Farmer, E. E., Almeras, E. & Krishnamurthy, V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6, 372–378 (2003)
Liechti, R. & Farmer, E. E. The jasmonate pathway. Science 296, 1649–1650 (2002)
Pauwels, L., Inze, D. & Goossens, A. Jasmonate-inducible gene: what does it mean? Trends Plant Sci. 14, 87–91 (2009)
Yan, J. et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236 (2009)
Fonseca, S. et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chem. Biol. 5, 344–350 (2009)
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y. & Howe, G. A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. USA 105, 7100–7105 (2008)
Thines, B. et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 (2007)
Xie, D.-X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998)
Yan, Y. et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483 (2007)
Chen, R. et al. The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916 (2012)
Fernandez-Calvo, P. et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011)
Cevik, V. et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis . Plant Physiol. 160, 541–555 (2012)
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405 (2010)
Melcher, K. The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional proteins. J. Mol. Biol. 301, 1097–1112 (2000)
Sun, X., Rikkerink, E. H., Jones, W. T. & Uversky, V. N. Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25, 38–55 (2013)
Triezenberg, S. J. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5, 190–196 (1995)
Smolen, G. A., Pawlowski, L., Wilensky, S. E. & Bender, J. Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis . Genetics 161, 1235–1246 (2002)
Figueroa, P. & Browse, J. The Arabidopsis JAZ2 promoter contains a G-Box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 53, 330–343 (2012)
Melotto, M. et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988 (2008)
Chung, H. S. et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622 (2010)
Kazan, K. & Manners, J. M. MYC2: the master in action. Mol. Plant 6, 686–703 (2013)
Kidd, B. N. et al. The Mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis . Plant Cell 21, 2237–2252 (2009)
Santner, A. & Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078 (2009)
Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010)
Doublie, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997)
Melcher, K. et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608 (2009)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Bailey, S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Ke, J. et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nature Struct. Mol. Biol. 20, 1377–1382 (2013)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Suino, K. et al. The nuclear xenobiotic receptor CAR; structural determinants of constitutive activation and heterodimerization. Mol. Cell 16, 893–905 (2004)
Xu, H. E. et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature 415, 813–817 (2002)
Withers, J. et al. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl Acad. Sci. USA 109, 20148–20153 (2012)
Yang, D. L. et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl Acad. Sci. USA 109, E1192–E1200 (2012)
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572 (2007)
Yao, J., Withers, J. & He, S. Y. Pseudomonas syringae infection assays in Arabidopsis . Methods Mol. Biol. 1011, 63–81 (2013)
Chalmers, M. J. et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006)
Pascal, B. D. et al. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 (2012)
Acknowledgements
This research is supported by the Gordon and Betty Moore Foundation (GBMF3037, to S.Y.H.), the China Scholarship Council (to F.Z.), Van Andel Research Institute (to H.E.X. and K.M.), the National Institutes of Health (R01 GM102545 to K.M. and R01AI060761 to S.Y.H.), and the Department of Energy (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science Grant DE–FG02–91ER20021 (to S.Y.H.). We thank S. Grant for administrative support and staff members of the Life Science Collaborative Access Team of the Advanced Photon Source for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). Use of the Advanced Photon Source was supported by the Office of Science of the US Department of Energy, under contract number DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank G. Howe and K. Aung for reading the manuscript.
Author information
Authors and Affiliations
Contributions
H.E.X., K.M., M.Z. and S.Y.H. conceived the project and designed experiments, F.Z., J.Y., J.K., L.Z., V.Q.L., X.F.X., X.E.Z., J.C., J.B. and P.R.G. performed and/or interpreted experiments. K.M., F.Z., J.Y. and S.Y.H. wrote the manuscript with support from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
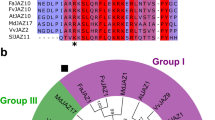
Extended Data Figure 1 Mapping of the JAZ9–MYC3 interface.
a, b, Yeast two-hybrid analysis of the interaction between LexA–JAZ9 and BD42(AD)–MYC3 constructs. Simplified diagrams of MYC3 (a) and JAZ9 (b) proteins are shown on top. Blue yeast colonies indicate a positive interaction between two proteins. The experiment was repeated three times with same results. c, Sequence alignment of the Jas motif of the 12 A. thaliana JAZ proteins. The N-terminal five amino acids that are unwound in the crystal structure of the COI1–ligand–JAZ co-receptor complex23 are indicated by a red line on top of JasJAZ1. Asterisks denote amino acids conserved in all of the sequences, colons denote similar amino acids. d, Interaction between purified His6Sumo–MYC3 N-terminal proteins and biotinylated Jas motif peptides by AlphaScreen luminescence proximity assay (n = 3 technical replicates; error bars, s.d.). ***Significant differences (P < 0.001) compared with the no-Jas control by Student’s t-test. The experiment was repeated three times with similar results.
Extended Data Figure 2 Interactions of MYC2 and MYC4 with Jas motifs of JAZ8, JAZ9 and JAZ12.
a, Yeast two-hybrid assays between MYC N-terminal proteins and full-length JAZ9. The experiment was repeated three times with same results. b, AlphaScreen assay between His6Sumo-tagged MYC N-terminal proteins and biotinylated JAZ peptides (n = 3 technical replicates; error bars, s.d.). ***Significant differences (P < 0.001) compared with no-Jas control by Student’s t-test. The experiment was repeated three times with similar results.
Extended Data Figure 3 Arrangement of secondary structure elements in MYC3(5–242).
a, Rainbow colour scheme of MYC3(5–242) in two orientations, from blue (N terminus) to red (C terminus). b, Secondary structure elements overlaid on the sequence alignment of MYC2, MYC3 and MYC4 N-terminal proteins. Note that α1′ (solid line) is a loop with partial helix character and is connected to α1 by a 90° kink.
Extended Data Figure 4 Surface accessibility and structural dynamics of MYC3(5–242) revealed by HDX.
a, HDX heat map of MYC3(5–242). The colour bar indicates the percentage deuterium exchange. Three experimental repeats were performed for each HDX time point. b, HDX heat map overlaid onto the MYC3(5–242) apo structure. Peptides corresponding to the JID helix were not resolved (no HDX information, grey colour), preventing a definitive assessment of the dynamics of the JID helix in solution.
Extended Data Figure 5 B-factor presentations of the four crystal structures.
The B-factor indicates the dynamic mobilities of different resolved parts within the structure. The thicker the lines and the warmer the colour, the higher is the mobility. Other than two linker regions (linker), the three helices that can occupy the JID helix have the highest B-factors in all four structures. The difficulties in crystallizing the MYC3(5–242)–Jas22JAZ9 complex are therefore probably caused by its high conformational flexibility due to the presence of all three dynamic helices as well as the unfolding of the α1′/α1 helix. Covalent fusion to MYC3(5–242) probably stabilizes the conformational flexibility of Jas22JAZ9 and the complex. Note that the presence of the α1′/α1 helix does not interfere with the ability of MYC3 to bind the JAZ peptide (compare MYC3(5–242) and MYC3(44–238) in Fig. 5b).
Extended Data Figure 6 Effects of mutations in MYC3 on the interactions between JAZ and MYC3 proteins in yeast two-hybrid assays.
The development of blue yeast colonies indicates the positive interaction between two proteins. The experiment was repeated three times with same results.
Extended Data Figure 7 MYC3(44–238) in complex with Jas22JAZ1 and Jas22JAZ9 and phenotypes of the Arabidopsis med25 mutant.
a, b, MYC3(44–238) in complex with Jas22JAZ1(amino acids 200–221; grey) overlaid with MYC3(44–238) in complex with Jas22JAZ9(amino acids 218–239; pink). c, Arabidopsis med25 mutant (pft1-2) plants are less susceptible to P. syringae pathovar tomato (Pst) DC3000 than Arabidopsis wild-type (Col-0) plants. Disease symptoms (chlorotic lesions; upper panel) and bacterial population (lower panel) of Arabidopsis wild-type and med25 mutant (pft1-2) plants 3 days after dip-inoculation with Pst DC3000 at 1 × 108 colony-forming units per millilitre (n = 4 biological replicates; error bars, s.e.m.). ***Significant difference (P < 0.001) in bacteria population between Col-0 and med25 mutant plants, as determined by two-tailed t-test. The experiment was repeated five times with similar results and the images presented are representative of five repeats. d, The med25 mutant plants are less sensitive to jasmonate-induced root growth inhibition than wild-type plants. A representative picture (upper panel) and percentages of root growth inhibition (lower panel) of 10-day-old wild-type and med25 mutant (pft1-2) Arabidopsis seedlings after treatment with 0.1% DMSO (control), 1 μM, 3 μM or 10 μM MeJA (n = 15 biological replicates; error bars, s.e.m.). Triple asterisks (***) with different colours indicate the significant differences (P < 0.001) between Col-0 and the med25 (pft1-2) mutant with the same concentration of MeJA treatment, as determined by two-way ANOVA with Bonferroni post-test. The experiment was repeated four times with similar results and the images presented are representative of four repeats.
Extended Data Figure 8 A simplified diagram of the core components of the jasmonate signalling cascade.
a, In the resting stage, jasmonate response gene expression is restrained by a family of JAZ transcriptional repressors. JAZ repressors bind and inhibit the MYC family of transcription factors through (1) direct inhibition and (2) recruiting TOPLESS (TPL) co-repressors either directly or through the NINJA adaptor. TPL in turn recruits histone deacetylases/methyltransferases (not shown) to repress gene expression through chromatin remodelling. b, In response to stress or developmental cues, plants synthesize jasmonate–Ile, which serves as molecular glue to facilitate the formation of a co-receptor complex between JAZ and COI1. The formation of the COI1–JAZ co-receptor complex leads to ubiquitination and proteasome-dependent degradation of JAZ repressors. c, JAZ-free MYCs interact with the MED25 subunit of the Mediator complex and recruit RNA polymerase II (not shown) to the promoters of jasmonate-responsive genes. Components examined in this study are coloured.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-2. (PDF 289 kb)
Conformational changes in MYC3 N-terminal domain (aa 5-242) upon JasJAZ9 peptide binding.
All the structures are shown in cartoon representation. MYC3 N-terminal domain structure is colored in cyan for JID domain, green for TAD domain, red for the partial α1 helix and yellow for the rest of the protein. JasJAZ9 peptide is colored in pink. Although the α1’/α1 helices are well-resolved in the apo MYC3 structure, only partial α1 helix is shown since the rest of the α1 helix is disordered in the MYC3(5-242)–JasJAZ9 complex structure. The movie was made using the Chimera program with the superimposed apo MYC3(5-242) and JasJAZ9 bound MYC3(5-242) structures. (AVI 663 kb)
Conformational changes in JasJAZ9 peptide transitioning from MYC3-bound state to COI1-bound state
All the structures are shown in cartoon representation with COI1 colored in blue and JasJAZ9 peptide colored in pink. JA-Ile is shown as a space-filling model. The starting conformation of JasJAZ9 peptide is shown as an α-helix based on JasJAZ9- bound MYC3 complex structure. JasJAZ9 peptide undergoes conformational change at the N terminus upon binding to COI1 and JA-Ile. The movie was made using the Chimera program with the superimposed JasJAZ9-bound MYC3 (this study) and JasJAZ1-bound COI123 structures. (AVI 1290 kb)
Rights and permissions
About this article
Cite this article
Zhang, F., Yao, J., Ke, J. et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273 (2015). https://doi.org/10.1038/nature14661
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14661
This article is cited by
-
The N-terminal α2 helix element is critical for the activity of the rice transcription factor MYC2
Plant Molecular Biology (2024)
-
The transcription factor GhWRKY70 from gossypium hirsutum enhances resistance to verticillium wilt via the jasmonic acid pathway
BMC Plant Biology (2023)
-
NLR surveillance of pathogen interference with hormone receptors induces immunity
Nature (2023)
-
Genome-wide analysis of JAZ family genes expression patterns during fig (Ficus carica L.) fruit development and in response to hormone treatment
BMC Genomics (2022)
-
Jasmonates and Histone deacetylase 6 activate Arabidopsis genome-wide histone acetylation and methylation during the early acute stress response
BMC Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.