Abstract
Abnormal accumulation of triglycerides in the liver, caused in part by increased de novo lipogenesis, results in non-alcoholic fatty liver disease and insulin resistance1,2. Sterol regulatory element-binding protein 1 (SREBP1), an important transcriptional regulator of lipogenesis, is synthesized as an inactive precursor that binds to the endoplasmic reticulum (ER). In response to insulin signalling, SREBP1 is transported from the ER to the Golgi in a COPII-dependent manner, processed by proteases in the Golgi, and then shuttled to the nucleus to induce lipogenic gene expression3,4,5; however, the mechanisms underlying enhanced SREBP1 activity in insulin-resistant obesity and diabetes remain unclear. Here we show in mice that CREB regulated transcription coactivator 2 (CRTC2)6 functions as a mediator of mTOR7 signalling to modulate COPII-dependent SREBP1 processing. CRTC2 competes with Sec23A, a subunit of the COPII complex8, to interact with Sec31A, another COPII subunit, thus disrupting SREBP1 transport. During feeding, mTOR phosphorylates CRTC2 and attenuates its inhibitory effect on COPII-dependent SREBP1 maturation. As hepatic overexpression of an mTOR-defective CRTC2 mutant in obese mice improved the lipogenic program and insulin sensitivity, these results demonstrate how the transcriptional coactivator CRTC2 regulates mTOR-mediated lipid homeostasis in the fed state and in obesity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Samuel, V. T. & Shulman, G. I. Mechanisms for insulin resistance: common threads and missing links. Cell 148, 852–871 (2012)
Perry, R. J., Samuel, V. T., Petersen, K. F. & Shulman, G. I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 510, 84–91 (2014)
Yellaturu, C. R. et al. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J. Biol. Chem. 284, 7518–7532 (2009)
Goldstein, J. L., DeBose-Boyd, R. A. & Brown, M. S. Protein sensors for membrane sterols. Cell 124, 35–46 (2006)
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002)
Altarejos, J. Y. & Montminy, M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature Rev. Mol. Cell Biol. 12, 141–151 (2011)
Lamming, D. W. & Sabatini, D. M. A Central role for mTOR in lipid homeostasis. Cell Metab. 18, 465–469 (2013)
Zanetti, G., Pahuja, K. B., Studer, S., Shim, S. & Schekman, R. COPII and the regulation of protein sorting in mammals. Nature Cell Biol. 14, 20–28 (2012)
Brown, M. S. & Goldstein, J. L. Selective versus total insulin resistance: a pathogenic paradox. Cell Metab. 7, 95–96 (2008)
Lin, H. V. & Accili, D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 14, 9–19 (2011)
Strable, M. S. & Ntambi, J. M. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit. Rev. Biochem. Mol. Biol. 45, 199–214 (2010)
Wang, Y. et al. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc. Natl Acad. Sci. USA 107, 3087–3092 (2010)
Song, Y. et al. CRTC3 links catecholamine signalling to energy balance. Nature 468, 933–939 (2010)
Altarejos, J. Y. et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nature Med. 14, 1112–1117 (2008)
Wang, Y. et al. Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature 485, 128–132 (2012)
Wang, Y., Vera, L., Fischer, W. H. & Montminy, M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 460, 534–537 (2009)
Bi, X., Mancias, J. D. & Goldberg, J. Insights into COPII coat nucleation from the structure of Sec23˙Sar1 complexed with the active fragment of Sec31. Dev. Cell 13, 635–645 (2007)
Peterson, T. R. et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011)
Li, S., Brown, M. S. & Goldstein, J. L. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl Acad. Sci. USA 107, 3441–3446 (2010)
Porstmann, T. et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236 (2008)
Yecies, J. L. et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 14, 21–32 (2011)
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011)
Khamzina, L., Veilleux, A., Bergeron, S. & Marette, A. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology 146, 1473–1481 (2005)
Um, S. H. et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 (2004)
Shimomura, I. et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell 6, 77–86 (2000)
Zhao, X. et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 122, 2417–2427 (2012)
Li, P. et al. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPAR activity and insulin sensitivity. Cell 147, 815–826 (2011)
Nohturfft, A., Yabe, D., Goldstein, J. L., Brown, M. S. & Espenshade, P. J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102, 315–323 (2000)
Schindler, A. J. & Schekman, R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc. Natl Acad. Sci. USA 106, 17775–17780 (2009)
Acknowledgements
We thank M. Montminy, W. E. Balch, L. Yu, S. Fu, P. Li, J. Wu and D. Li for discussion, and X. Liu and P. Li for technical help with lipogenesis analyses and clamp studies, respectively. This work was supported by grants from 1000 Talents Program for Young Scholars and the National Science Foundation of China (31322027, 31321003 and 31471127) to Y.W.
Author information
Authors and Affiliations
Contributions
J.H., E.L. and Y.W. designed the study and analysed the data. J.H., E.L., L.C., Y.Z., F.W. and Y.W. performed the experiments. J.L. and H.D. carried out mass spectrometry analysis. J.H. and Y.W. wrote the paper. All authors reviewed and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
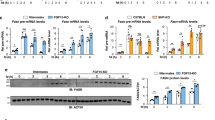
Extended Data Figure 1 Profiling of cholesterol, gene expression, protein and circulating insulin levels in Crtc2+/+ and Crtc2−/− mice.
a–d, Hepatic cholesterol levels (a), qPCR results for expression of genes involved in lipogenic regulation, lipid transport, fatty acid oxidation and triglyceride synthesis (b), plasma insulin level (c), and immunoblots showing hepatic amounts of full-length, inactive SREBP2 (flSREBP2) and cleaved, active SREBP2 (nSREBP2), phospho-CREB (pCREB), CREB, and subunits of the COPII complex (Sec12, Sar1, Sec23A, Sec24A, Sec13 and Sec31A) (d) in Crtc2+/+ and Crtc2−/− mice fed with a regular diet (RD) or a high-fat diet (HFD) for 18 weeks. Data are shown as mean ± s.e.m. *P < 0.01, n = 10.
Extended Data Figure 2 Validation of Srebp1 knockdown in mice.
a–c, Hepatic triglycerides (TGs) (a), immunoblots (b) and qPCR results for lipogenic gene expression (c) showing the effect of Srebp1 RNAi in liver extracts of fed mice. Data are shown as mean ± s.e.m. *P < 0.01, n = 10. US, unspecific.
Extended Data Figure 3 Effect of CRTC2 and its mutants on gluconeogenic and lipogenic gene expression.
a, Cellular localization of CRTC2 and its mutants CRTC2(ΔTAD) (amino acids 1–630) and CRTC2(ΔTAD/AA) (amino acids 1–630 with double alanine mutations at Ser171 and Ser275) in mouse primary hepatocytes. FSK, forskolin. Scale bar, 10 μm. b, Effect of CRTC2 and its mutants on Cre-luc activity in mouse primary hepatocytes treated with or without 10 μM FSK for 4 h. Data are shown as mean ± s.e.m. *P < 0.01, n = 6. c, d, Effect of CRTC2 and its mutants on lipogenic gene (Fasn, Scd1, Acaca) expression (c) and plasma insulin level (d) in fed mice. Data are shown as mean ± s.e.m. *P < 0.01, n = 8. NS, no significant statistical difference.
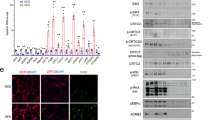
Extended Data Figure 4 Association of CRTC2 with Sec31A.
a, Immunostaining showing relative co-localization of CRTC2 with an endoplasmic reticulum (ER) marker (KDEL) in CRL-2189 cells. Scale bar, 10 μm. b, c, Strategy to purify CRTC2-interacting proteins (b), and the peptides identified from Sec31A and Sec13 (c) by MS analysis of immunoprecipitates prepared with anti-CRTC2 antibody from CRL-2189 ER fractions. d, Co-immunoprecipitation assay showing amounts of Flag-tagged Sec13 or Sec31A recovered from immunoprecipitates of endogenous CRTC2 in HEK293T cells. e, Deletion analysis of regions in CRTC2 required for the CRTC2–Sec31A interaction. Interaction-competent CRTC2 peptides are indicated by (+) in each schematic. f, g, Cellular localization of the tryptophan-to-alanine mutant of CRTC2 (W143A) and its effect on Cre-luc activity in HEK293T cells. Scale bars, 10 μm. Data are shown as mean ± s.e.m. *P < 0.01, n = 6. NS, no significant statistical difference. h, Co-immunoprecipitation assay showing amounts of Flag-tagged CRTC2 and YFP-tagged Sec23A recovered from immunoprecipitates of HA-tagged Sec31A in HEK293T cells.
Extended Data Figure 5 Modulation of COPII-dependent SREBP1 activity by CRTC2.
a, Immunostaining showing the effect of Crtc2 RNAi on the cellular localization of Sec31A. Scale bars, 10 μm. b, c, Effect of wild-type (WT) and Sec31A-interaction-defective (W143A) CRTC2 on lipogenic gene expression (b) and plasma insulin level (c) in fed mice. Data are shown as mean ± s.e.m. *P < 0.01, n = 8. NS, no significant statistical difference.
Extended Data Figure 6 Characterization of CRTC2 phosphorylation site(s) by mTOR.
a, Immunoblots showing co-immunoprecipitation of CRTC2 and Sec23A with Sec31A in mouse primary hepatocytes in response to insulin and/or torin1 treatment. Mouse primary hepatocytes were incubated with 250 nM torin1 or control vehicle for 1 h before 30 min insulin (100 nM) stimulation. Phospho-S6K (pS6K), total S6K, phospho-AKT (pAKT), total AKT and phospho-CRTC2 (Ser136) levels are also indicated. b, Immunoblots showing co-immunoprecipitation of CRTC2 and Sec23A with Sec31A in mouse primary hepatocytes in response to amino acids and/or torin1 treatment. Mouse primary hepatocytes incubated with amino-acid-free MEM for 3 h were exposed to 250 nM torin1 or control vehicle for another 1 h, then treated with amino acids for 30 min. c, Phospho-peptides of Flag-tagged CRTC2 identified by MS analysis of immunoprecipitates prepared with anti-Flag from HEK293T cells treated with 100 nM rapamycin for 1 h (Rap+) or not (Rap−). Serine 136 was phosphorylated (Yes) in the absence of Rap treatment (Rap−) and dephosphorylated (No) in the presence of Rap (Rap+). d, Co-immunoprecipitation assay showing the association between Flag-tagged CRTC2 and Myc-tagged mTOR in HEK293T cells. e, Effect of the mTOR inhibitors Rap and torin1 on CRTC2 phosphorylation. Mouse primary hepatocytes were pretreated with vehicle (Veh), 100 nM Rap, or 250 nM torin1 for 1 h before 100 nM insulin stimulation for 30 min. f, g, Cellular localization of the phosphorylation-defective CRTC2 mutant (S136A) (f) and its effect on Cre-luc activity (g) in mouse primary hepatocytes. Scale bars, 10 μm. Data are shown as mean ± s.e.m. *P < 0.01, n = 6. NS, no significant statistical difference.
Extended Data Figure 7 Effect of CRTC2(S136A) on SREBP1 maturation, lipin1 localization and circulating insulin level.
a, b, Effect of wild-type or CRTC2(S136A) on lipogenic gene expression in liver (a) and plasma insulin level (b) of fasted (3 h) and refed (1 h after 3 h fasting) mice. c–e, Effect of CRTC2(S136A) and torin1 treatment on SREBP1 maturation (c), hepatic triglycerides (d) and lipin1 localization in mouse primary hepatocytes (e). Torin1 (20 mg kg−1) was intraperitoneally injected 6 h before livers were harvested. For lipin1 localization, mouse primary hepatocytes were treated with vehicle (Torin1−) or 250 nM torin1 (Torin1+) for 4 h. Scale bars, 10 μm. Data are shown as mean ± s.e.m. *P < 0.01, n = 8. NS, no significant statistical difference.
Extended Data Figure 8 Enhanced SREBP1 activation, triglyceride levels and branched-chain amino acid levels in obese mice.
a–c, Immunoblots showing relative amounts and/or phosphorylation status of SREBP1, SREBP2, SCAP, mTOR, S6K, CRTC2, AKT and COPII subunits in fed lean and db/db mice (a), ob/ob mice (b), and relative amounts of SREBP2, SCAP and COPII subunits in HFD-fed mice (c). d–g, Hepatic triglyceride amounts and branched-chain amino acid (BCAA) levels in liver extracts from lean, db/db, ob/ob and HFD-fed mice in the fed state. Data are shown as mean ± s.e.m. *P < 0.01, n = 10.
Extended Data Figure 9 Improved insulin sensitivity in HFD-fed mice in the presence of CRTC2(ΔTAD/S136A).
a–k, Effect of the mTOR-defective mutant CRTC2(ΔTAD/S136A) on metabolic parameters (a), including body weight, relative fat mass, food intake, plasma alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activity, plasma cholesterol, plasma triglycerides, plasma insulin and blood glucose; energy expenditure (b); lipogenic gene expression (c); glucose tolerance (d); insulin tolerance (e); pyruvate tolerance (f); hepatic glucose production (HGP; g); insulin-stimulated glucose disposal rate (IS-DGR; h); percentage of free fatty acid (FFA) suppression (i); pAKT level in skeletal muscle (j); and pAKT level in epididymal white adipose tissue (k) from mice fed on a HFD for 18 weeks. Data are shown as mean ± s.e.m. *P < 0.01, **P < 0.05, n = 8 (a–f), n = 6 (g–i).
Rights and permissions
About this article
Cite this article
Han, J., Li, E., Chen, L. et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 524, 243–246 (2015). https://doi.org/10.1038/nature14557
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14557
This article is cited by
-
Unveiling the dark side of glucose-regulated protein 78 (GRP78) in cancers and other human pathology: a systematic review
Molecular Medicine (2023)
-
The CRTC-1 transcriptional domain is required for COMPASS complex-mediated longevity in C. elegans
Nature Aging (2023)
-
Famsin, a novel gut-secreted hormone, contributes to metabolic adaptations to fasting via binding to its receptor OLFR796
Cell Research (2023)
-
Regulation and targeting of SREBP-1 in hepatocellular carcinoma
Cancer and Metastasis Reviews (2023)
-
Biological functions of CRTC2 and its role in metabolism-related diseases
Journal of Cell Communication and Signaling (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.