Abstract
The endoplasmic reticulum (ER) is the largest intracellular endomembrane system, enabling protein and lipid synthesis, ion homeostasis, quality control of newly synthesized proteins and organelle communication1. Constant ER turnover and modulation is needed to meet different cellular requirements and autophagy has an important role in this process2,3,4,5,6,7,8. However, its underlying regulatory mechanisms remain unexplained. Here we show that members of the FAM134 reticulon protein family are ER-resident receptors that bind to autophagy modifiers LC3 and GABARAP, and facilitate ER degradation by autophagy (‘ER-phagy’). Downregulation of FAM134B protein in human cells causes an expansion of the ER, while FAM134B overexpression results in ER fragmentation and lysosomal degradation. Mutant FAM134B proteins that cause sensory neuropathy in humans9 are unable to act as ER-phagy receptors. Consistently, disruption of Fam134b in mice causes expansion of the ER, inhibits ER turnover, sensitizes cells to stress-induced apoptotic cell death and leads to degeneration of sensory neurons. Therefore, selective ER-phagy via FAM134 proteins is indispensable for mammalian cell homeostasis and controls ER morphology and turnover in mice and humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Borgese, N., Francolini, M. & Snapp, E. Endoplasmic reticulum architecture: structures in flux. Curr. Opin. Cell Biol. 18, 358–364 (2006)
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011)
Schuck, S., Prinz, W. A., Thorn, K. S., Voss, C. & Walter, P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187, 525–536 (2009)
Maiuolo, J., Bulotta, S., Verderio, C., Benfante, R. & Borgese, N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc. Natl Acad. Sci. USA 108, 7832–7837 (2011)
Hamasaki, M., Noda, T., Baba, M. & Ohsumi, Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6, 56–65 (2005)
Bernales, S., McDonald, K. L. & Walter, P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423 (2006)
Tasdemir, E. et al. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle 6, 2263–2267 (2007)
Yorimitsu, T. & Klionsky, D. J. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 17, 279–285 (2007)
Kurth, I. et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nature Genet. 41, 1179–1181 (2009)
Stolz, A., Ernst, A. & Dikic, I. Cargo recognition and trafficking in selective autophagy. Nature Cell Biol. 16, 495–501 (2014)
Weidberg, H., Shvets, E. & Elazar, Z. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 (2011)
Rogov, V., Dotsch, V., Johansen, T. & Kirkin, V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178 (2014)
Kirkin, V. et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009)
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011)
Rogov, V. V. et al. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem. J. 454, 459–466 (2013)
Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. & Rapoport, T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586 (2006)
Shibata, Y. et al. Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788 (2010)
Schuck, S., Gallagher, C. M. & Walter, P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127, 4078–4088 (2014)
Cebollero, E., Reggiori, F. & Kraft, C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int. J. Cell Biol. 2012, 182834 (2012)
Beetz, C. et al. A spastic paraplegia mouse model reveals REEP1-dependent ER shaping. J. Clin. Invest. 123, 4273–4282 (2013)
Hübner, C. A. & Kurth, I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain 137, 3109–3121 (2014)
Renvoisé, B. & Blackstone, C. Emerging themes of ER organization in the development and maintenance of axons. Curr. Opin. Neurobiol. 20, 531–537 (2010)
Spoerri, P. E., Dresp, W. & Heyder, E. A simple embedding technique for monolayer neuronal cultures grown in plastic flasks. Acta Anat. 107, 221–223 (1980)
Mao, K., Wang, K., Liu, X. & Klionsky, D. J. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell 26, 9–18 (2013)
Mochida, K. et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Naturehttp://dx.doi.org/10.1038/nature14506 (2015)
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010)
Altan-Bonnet, N. et al. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Mol. Biol. Cell 17, 990–1005 (2006)
GrandPré, T., Nakamura, F., Vartanian, T. & Strittmatter, S. M. Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature 403, 439–444 (2000)
Klopfenstein, D. R. et al. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J. Cell Biol. 153, 1287–1300 (2001)
Stewart, S. A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003)
Popovic, D. et al. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32, 1733–1744 (2012)
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004)
Yang, M., Ellenberg, J., Bonifacino, J. S. & Weissman, A. M. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272, 1970–1975 (1997)
Chan, E. Y., Kir, S. & Tooze, S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 (2007)
Verheije, M. H. et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4, e1000088 (2008)
Slot, J. W. & Geuze, H. J. Cryosectioning and immunolabeling. Nature Protocols 2, 2480–2491 (2007)
Khundadze, M. et al. A hereditary spastic paraplegia mouse model supports a role of ZFYVE26/SPASTIZIN for the endolysosomal system. PLoS Genet. 9, e1003988 (2013)
Le Bars, D., Gozariu, M. & Cadden, S. W. Animal models of nociception. Pharmacol. Rev. 53, 597–652 (2001)
Acknowledgements
We would like to thank S. Horwitz, K. Rajalingam, C. Behrends and J. Lippincott-Schwartz for cell lines and vectors, N. Mizushima for Atg5–/– and control immortalized MEFs, H.-P. Hauri and H. Farhan for vectors, and S. Gießelmann and K. Schorr for excellent technical assistance. We acknowledge D. McEwan, D. Hoeller, D. Popovic and K. Koch for critical reading of the manuscript and valuable insights. We also thank M. M. Kessels for support. This work was supported by grants from the Deutsche Forschungsgemeinschaft to I.D. (DI 931/3-1), I.K. (KU 1587/2-1, KU 1587/3-1, KU 1587/4-1), C.A.H. (HU 800/5-1, RTG 1715, HU 800/6-1, HU 800/7-1), B.Q. (QU116/6-2, RTG1715), J.W. (WE1406/13-1), the Cluster of Excellence ‘Macromolecular Complexes’ of the Goethe University Frankfurt (EXC115), LOEWE grant Ub-Net and LOEWE Centrum for Gene and Cell therapy Frankfurt and the European Research Council/ERC grant agreement number (250241-LineUb) to I.D. F.R. is supported by ECHO (700.59.003), ALW Open Program (821.02.017 and 822.02.014), DFG-NWO cooperation (DN82-303) and ZonMW VICI (016.130.606) grants. P.G. is supported by the 7.FP, COFUND, Goethe International Postdoc Programme GO-IN, No. 291776.
Author information
Authors and Affiliations
Contributions
A.K. performed biochemical analyses, immunofluorescence and cellular localization, functional analysis and contributed to interpretation of data and manuscript writing and preparation. T.H. characterized Fam134b–/– mice, carried out FAM134B topology analysis and contributed to manuscript preparation. M.Mar. performed transmission electron microscopy of cells and neurons in culture. P.G. performed apoptosis and autophagy analysis, and contributed to manuscript preparation and writing. A.K.H. generated the Fam134b–/– mouse model and was involved in mouse phenotyping. M.A. performed crystal structure assay. L.L. performed the electrophysiological analysis of Fam134b–/– mice. S.N., I. Ka. and J.W. performed transmission electron microscopy on murine tissues. A.S. performed fractionation and autophagy flux experiments. M.Mau. carried out the assay for the turnover of long-lived proteins. N.K. performed liposome assays, B.Q. supervised liposome assays. F.R., I.Ku., C.A.H. and I.D. designed the study, analysed data and wrote the manuscript. I.Ku., C.A.H. and I.D. contributed equally to the study. All the authors discussed the results and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 FAM134 proteins bind to GST-LC3-like modifiers.
a, A549 cell lysates were added to beads with various immobilized GST fusion proteins (GST, GST–LC3A, GST–LC3B, GST–GABARAP, GST–GABARAP–L1), followed by western blotting using an antibody against FAM134B. b, Co-immunoprecipitation between Fam134b and Lc3b in wild-type (WT) mouse embryonic fibroblasts (MEFs). MEFs isolated from Fam134b-knockout (KO) mice served as control. c, Interaction of FAM134A and FAM134C with LC3-like modifiers. d, FAM134B proteins lacking the reticulon domain (Δret), disease-associated truncating FAM134B variants Q145X and S309X, and FAM134B with a mutated LIR-motif (mutLIR) fail to bind GST–LC3-like modifiers (LC3B) in contrast to wild-type FAM134B and FAM134B lacking the coiled-coil domain (ΔCC). e, Crystal structure of a FAM134B-LIR peptide interacting with LC3A. Ribbon and surface representation model of the FAM134B-LIR–LC3A interaction and crystallographic symmetry related molecule. (LIR, magenta; LC3A, yellow). 2Fo − Fc electron density (blue mesh) of FAM134B-LIR (amino acids 453–459), contoured at 1σ and with ball-and-stick model.
Extended Data Figure 2 Subcellular localization and topology of endogenous FAM134B.
a, The post-nuclear fraction of wild-type MEF lysate was loaded on a linear Iodixanol gradient (0–20%). Fractions (top to bottom) were subjected to western blot and analysed with antibodies directed against endogenous proteins as indicated. Images are from two different gels, while the exposure time is the same. Asterik indicates non-specific band. b, A549 cells and MEFs were transfected with a KDEL–RFP expression plasmid for 24 h. After fixation, endogenous FAM134B was detected. FAM134B co-localizes with KDEL–RFP. Representatives of five images are shown. Scale bar, 10 μm. c, HeLa and U2OS cells were fixed and stained for endogenous FAM134B and CLIMP-63. FAM134B co-localizes with CLIMP-63. Representatives of five images are shown. Scale bar, 10 μm. d, FAM134B topology analysis. COS-7 cells transiently overexpressing C-terminal tagged FAM134B (FAM134B–eGFP) or N-terminal tagged FAM134B (eGFP–FAM134B) were subjected to fluorescence protein protection (FPP) assay. COS-7 cells transfected with plasmids encoding the luminal ER peptide RFP–KDEL and C-terminal RFP-tagged CD3 (CD3–RFP) served as controls. RFP–KDEL, which localized to the ER lumen, shows fluorescence protein protection upon trypsin administration following digitonin treatment. By contrast, according to the known topology of CD3–RFP the RFP-tag faces the cytosol and as a consequence trypsin treatment abolishes protein fluorescence. Scale bar, 10 μm. e, C-terminal RFP-tagged CD3 (CD3–RFP) served as control (RFP-tag faces the cytosol). A N-terminal tagged FAM134B variant was also subjected to the FPP assay. A strong decrease in fluorescence was observed for CD3–RFP, FAM134B–eGFP and eGFP–FAM134B after sequenced digitonin and trypsin treatment, but not for RFP–KDEL (n = number of cells, error bars indicate s.e.m.).
Extended Data Figure 3 FAM134B is a membrane-shaping protein that remodels lipid bilayers.
a, Liposome co-floatation assays. Proteins were detected using anti-GST antibodies in immunoblots of sucrose gradient fractions 1 (top) to 6 (bottom). GST–FAM134B, but not GST (Ctrl), floated with liposomes in fraction 2. Disease-related truncating mutations S309X and Q145X only partially floated with the liposome fraction. b, Representative transmission electron microscopy (TEM) images of freeze-fractured incubations of liposomes with recombinant GST-fusion proteins (GST–FAM134B; GST–FAM134B(Q145X); Ctrl, GST). Scale bar, 200 nm. c, Distribution of liposome diameters observed by TEM of freeze-fractured liposome incubations. Incubation with FAM134B and S309X leads to a pronounced increase in the relative numbers of smaller structures in comparison to control (GST) and Q145X. Inset, box plots of data presented in c; y axis is logarithmic. n.s., not significant; ***P < 0.001 one-way ANOVA, error bars indicate s.e.m. (n = 2,682 for Ctrl, n = 2,683 for FAM134B, n = 1,685 for Q145X, n = 1,612 for S309X, n = number of liposomes). Boxes contain 50% of the values; minimal, maximal, and median values are marked by vertical lines.
Extended Data Figure 4 Fam134b determines ER degradation through autophagy.
a, U2OS cells transiently co-transfected with 0.25 µg of GFP–SEC61 plasmid expressing full length or mutLIR FAM134B–HA at the indicated quantities (µg). Cell lysates were immunoblotted with antibodies against HA, GFP and vinculin. Black arrow-head indicates protein-degradation products. b, Wild-type (WT) and Atg5-knockout (KO) MEFs transiently expressing mCherry–eGFP–FAM134B were fixed and processed for immunofluorescence analysis. Cells with mCherry-positive and simultaneously GFP-negative punctae were counted in three independent experiments (biological replicates). Representative of 50 images is shown. Scale bar, 10 μm. c, d, Atg5-knockout and control MEFs were co-transfected with plasmids expressing full-length LIR (c) or mutLIR (d) FAM134B–HA and KDEL–RFP for 24 h, fixed and processed for immunofluorescence using antibodies against the HA tag and Lc3b. Overexpression of FAM134B–HA in wild-type but not in Atg5-knockout MEFs leads to the formation of punctae positive for both KDEL–RFP and Lc3b. Representatives of five images are shown. Scale bar, 10 μm. e, Quantification of cells displaying KDEL–RFP- and Lc3b-positive punctae shown in c and d. At least 50 cells per experiment from three independent experiments (biological replicates) were quantified (error bars indicate s.d.). f, Fam134b is stabilized in starved Atg5-knockout MEFs. Wild-type and Atg5-knockout MEFs were starved in EBSS for the indicated periods of time. Cell lysates were processed by SDS–PAGE and western blot using antibodies against Fam134b and vinculin.
Extended Data Figure 5 FAM134B knockdown causes ER expansions.
a, Ultrastructural analysis of FAM134B-depleted cells. Constructs expressing control shRNA or α-FAM134B shRNA were lentivirally delivered into U2OS cells. Cells were chemically fixed and embedded with Epon resin. Longitudinal and transversal sections and a part of the nuclear membrane is shown. FAM134B-depleted cells display ER expansions, particularly in the cell periphery. The membrane of the nucleus of FAM134B-knockdown cells also undergoes expansion. M, mitochondria; N, nucleus; PM, plasma membrane. Scale bars, 2 μm (left images) and 500 nm (middle and right images) (n = 150 cells). b, c, ER sheets are expanded in FAM134B-deficient cells. Constructs expressing control shRNA or α-FAM134B shRNA were lentivirally delivered in U2OS cells. After selection, cells were fixed, stained for CLIMP-63 and RTN4A and RTN4B and analysed by fluorescence microscopy. Representatives of five images are shown. Scale bar, 10 μm. d, CLIMP-63 and TRAPα levels in autophagy-deficient and FAM134B-depleted cells.
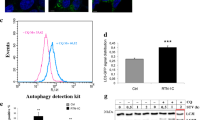
Extended Data Figure 6 FAM134B is not required for bulk autophagy and aggrephagy.
a, Fam134b+/+ and Fam134b–/– MEFs were starved in EBSS for 2 h, fixed and stained for Lc3b (using two different anti-Lc3b antibodies to enhance the signal). Representatives of five images are shown. Scale bar, 10 μm. b, Lc3b-positive punctae in 50 cells per experiment from three independent experiments were counted. *P < 0.0001, one-way ANOVA. c, Long-living proteins degradation assay. For autophagy induction, cells were starved with EBSS. Protein degradation was assessed in three independent experiments in triplicate. Error bars represent s.e.m. of three independent counts (three technical replicates, n = 20 cells per each replicate, P value is calculated using t-test, *P < 0.05). d, Transmission electron microscopy of Fam134b+/+ and Fam134b–/– cells. Autophagosomal/degradative compartments (that is, autophagosomes, autolysosomes and lysosomes) were counted. Quantification was performed by counting 20 cells in three different grids (three biological replicates). Error bars represent s.d., P value is calculated using t-test. e, Lc3b lipidation and p62 degradation was analysed in Fam134b+/+ and Fam134b–/– MEFs treated with 200 nM bafilomycin A1 for the indicated time. f, g, For autophagy induction, cells were starved with EBSS (f) or treated with the chemical Ku-0063794 (10 µM) (g) for the indicated periods of time. Bafilomycin A1 was added 1 h before the beginning of the treatment. h, Control and FAM134B-knockdown (shRNA-mediated) U2OS cells were starved in EBSS for 2 h, fixed and stained for LC3B (using two different anti-LC3B antibodies to enhance the signal). Representatives of five images are shown. Scale bar, 10 μm. i, LC3B-positive punctae in 150 cells per experiment from three independent experiments were counted. *P < 0.0001, Mann–Whitney U-test. j, LC3B lipidation and p62 degradation were analysed in control and FAM134B shRNA cells treated with 200 nM bafilomycin A1 for the indicated time. k, l, For autophagy induction, cells were starved with EBSS (k) or treated with the chemical Ku-0063794 (10 µM) (l) for the indicated periods of time. Bafilomycin A1 was added 1 h before the beginning of the treatment. m, Fam134b+/+ and Fam134b–/– MEFs were either treated with 5 µg ml−1 puromycin for 2 h or treated and subsequently washed and incubated in puromycin-free medium for 3 h, fixed and stained for ubiquitin (Ub) and p62. Representatives of five images are shown. Scale bar, 10 μm. n, The number of cells with Ub/p62 punctae before and after puromycin release per 100 cells per experiment was determined from three independent experiments (biological replicates). Error bars indicate s.d.
Extended Data Figure 7 FAM134B deficiency sensitizes cells to apoptosis.
a, b, Fam134b+/+ and Fam134b–/– MEFs were treated with 25 µM CCCP (carbonyl cyanide 3-chlorophenylhydrazone) (a) or starved in EBSS(b) for the time indicated. Cell lysates were processed by SDS–PAGE and western blot using antibodies against vinculin, Parp and Fam134b. c, Fam134b+/+ and Fam134b–/– MEFs were treated with 2 μg ml−1 tunicamycin (Tm) or 1 μM thapsigargin (Tg) for 12 h or left untreated. Cell lysates were processed by SDS–PAGE and western blot using antibodies against vinculin and Parp. d, FACS analysis for annexin V and propidium iodide (PI) in Fam134b+/+ and Fam134b–/– MEFs. Quantifications of annexin V/propidium iodide-positive cells after the indicated experimental settings (EBSS starvation for 8 h, treatment with 1 μM thapsigargin, 2 μg ml−1 tunicamycin, 25 μM CCCP for 12 h and 200 nM staurosporin (STS) for 6 h). Data are shown as mean ± s.d. of two independent biological replicates; for each biological replicate two experiments were performed and for each experiment 10,000 cells were analysed. **P < 0.01, one-way ANOVA. e, A549 cells stably expressing control and anti-FAM134B no. 1 and no. 2 shRNAs were either starved in EBSS or treated with 10 μM Ku-0063794, 200 ng ml−1 bafilomycin A1 (BafA), 1 μM thapsigargin or 2 μg ml−1 tunicamycin for 12 h or left untreated. Cell lysates were processed by SDS–PAGE and western blot using antibodies against vinculin and PARP. f, g, FAM134B knockdown induces apoptosis involving the mitochondrial pathway. A549 cells stably expressing control and anti-FAM134B no. 2 shRNAs were starved in EBSS for the time indicated. Cell lysates were processed by SDS–PAGE and western blot using antibodies against vinculin, PARP and caspase 8 (f) or caspase 9 (g). a–c, e–g, Filled arrowheads indicate processed PARP and caspase 8/9; empty arrowheads indicate unprocessed PARP and caspase 8/9.
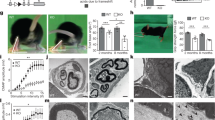
Extended Data Figure 8 Fam134b deficiency does not affect spinal cord motor neurons.
a, Western blot analysis of mouse embryonic tissue lysates with a Fam134b antibody. b, Transmission electron microscopy analysis of Fam134b+/+ and Fam134b–/– MEFs. Cells lacking Fam134b display an expanded ER. M, mitochondria. Scale bar, 1 μm; n = 150 cells. c, Compound muscle action potential (CMAP). CMAP latencies were recorded from tail nerves of wild-type (+/+) and knockout (−/−) mice at the ages indicated. No significant differences were observed between Fam134b+/+ and Fam134b−/− mice. One-way ANOVA, error bars indicate s.e.m.; n = 6 for +/+ and −/− at 6 months; n = 10 for +/+ and n = 13 for −/− at 12 months. n.s., not significant. d, Motor neurons in Nissl-stained thoracic spinal cord sections (10-µm thick) of Fam134b+/+ and Fam134b−/− mice. Scale bar, 20 µm. Representatives of seven images per genotype are shown. e, Quantification of motor neuron cell bodies in the ventral horn. Motor neuron number was unchanged in Fam134b+/+ and Fam134b−/− mice at an age of 12 and 20 months (n = 7 for +/+ and n = 7 for −/−). n.s., not significant, Student’s t-test, error bars indicate s.e.m. f, Normal ultrastructure of motor neurons in spinal cord sections of 12-month-old mice. Arrows in the middle and right panels highlight that there were no observed alterations in ER and Golgi architecture, respectively. Scale bar, 1 µm. Representatives of three images per genotype are shown.
Extended Data Figure 9 Ultrastructural analysis of dorsal root ganglia neurons in Fam134b−/− mice.
a–e, Ex vivo analysis of dorsal root ganglia (DRG) neurons. DRG neurons from 3-month-old Fam134b+/+ and Fam134b−/− littermates were cultured for 2 days before transmission electron microscopy using the flat embedding approach. a, b, The ultrastructural architecture of the peripheral ER as found in the DRG neuron cell body above or below the nucleus (a) and ER (b) adjacent to the nucleus of DRG neurons is shown. Scale bars: a, 500 nm; b, 200 nm. c, Representative examples of Golgi compartments. Scale bar, 200 nm. (n = 25 cells for a–c). d, e, Lateral cross-sections of the axons recognizable by their heavy myelination around the plasma membrane and their emptiness in organelles. Panel d shows a bundle of axons whereas panel e presents a single axon. e, lower panels, longitudinal cross-sections of neurites, which are typically packed with microtubules and ER. ER appears as thin black stripes in these images. ax, axon; ER, endoplasmic reticulum; G, Golgi; M, mitochondrium; mt, microtubule; my, myelin; N, nucleus, LD, lipid droplet. Scale bars: d, 1 µm; e, upper panels, 1 μm; lower panels, 500 nm. Representatives of 25 images per genotype are shown.
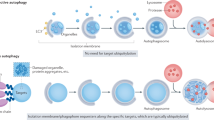
Extended Data Figure 10 Model of FAM134B function.
To drive ER-phagy, FAM134B clusters at the edges of cisternal ER. Local enrichment of FAM134B LIR leads to the recruitment of autophagosomal membranes and subsequent budding of ER-derived vesicles. Mature autophagosomes fuse with lysosomes leading to the degradation of enclosed ER fragments.
Supplementary information
Supplementary Information
This file contains the Western blot scans for figures 1, 3, 4 in the main paper and extended data figures 1, 2, 4, 5, 6, 7, 8. It also contains Supplementary Tables 1 and 2. (PDF 4662 kb)
Rights and permissions
About this article
Cite this article
Khaminets, A., Heinrich, T., Mari, M. et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358 (2015). https://doi.org/10.1038/nature14498
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14498
This article is cited by
-
Selective autophagy in cancer: mechanisms, therapeutic implications, and future perspectives
Molecular Cancer (2024)
-
Inhibition of OGFOD1 by FG4592 confers neuroprotection by activating unfolded protein response and autophagy after ischemic stroke
Journal of Translational Medicine (2024)
-
DDRGK1-mediated ER-phagy attenuates acute kidney injury through ER-stress and apoptosis
Cell Death & Disease (2024)
-
The Multiple Roles of Autophagy in Neural Function and Diseases
Neuroscience Bulletin (2024)
-
Proteome census upon nutrient stress reveals Golgiphagy membrane receptors
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.