Abstract
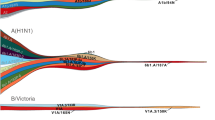
Understanding the spatiotemporal patterns of emergence and circulation of new human seasonal influenza virus variants is a key scientific and public health challenge. The global circulation patterns of influenza A/H3N2 viruses are well characterized1,2,3,4,5,6,7, but the patterns of A/H1N1 and B viruses have remained largely unexplored. Here we show that the global circulation patterns of A/H1N1 (up to 2009), B/Victoria, and B/Yamagata viruses differ substantially from those of A/H3N2 viruses, on the basis of analyses of 9,604 haemagglutinin sequences of human seasonal influenza viruses from 2000 to 2012. Whereas genetic variants of A/H3N2 viruses did not persist locally between epidemics and were reseeded from East and Southeast Asia, genetic variants of A/H1N1 and B viruses persisted across several seasons and exhibited complex global dynamics with East and Southeast Asia playing a limited role in disseminating new variants. The less frequent global movement of influenza A/H1N1 and B viruses coincided with slower rates of antigenic evolution, lower ages of infection, and smaller, less frequent epidemics compared to A/H3N2 viruses. Detailed epidemic models support differences in age of infection, combined with the less frequent travel of children, as probable drivers of the differences in the patterns of global circulation, suggesting a complex interaction between virus evolution, epidemiology, and human behaviour.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Russell, C. A. et al. The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 (2008).
Lemey, P. et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 10, e1003932 (2014).
Rambaut, A. et al. The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (2008).
Bedford, T., Cobey, S., Beerli, P. & Pascual, M. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog. 6, e1000918 (2010).
Chan, J., Holmes, A. & Rabadan, R. Network analysis of global influenza spread. PLOS Comput. Biol. 6, e1001005 (2010).
Nelson, M. I., Simonsen, L., Viboud, C., Miller, M. A. & Holmes, E. C. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 3, e131 (2007).
Nelson, M. I. et al. Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog. 2, e125 (2006).
Glezen, P. W., Schmier, J. K., Kuehn, C. M., Ryan, K. J. & Oxford, J. The burden of influenza B: a structured literature review. Am. J. Public Health 103, e43–e51 (2013).
Thompson, W. W. et al. Influenza-associated hospitalizations in the United States. J. Am. Med. Assoc. 292, 1333–1340 (2004).
Squires, R. B. et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir. Viruses 6, 404–416 (2012).
Chen, R. & Holmes, E. C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 66, 655–663 (2008).
Cox, N. J. & Bender, C. A. The molecular epidemiology of influenza viruses. Semin. Virol. 6, 359–370 (1995).
Bedford, T. et al. Integrating influenza antigenic dynamics with molecular evolution. eLife 3, e01914 (2014).
Bedford, T., Cobey, S. & Pascual, M. Strength and tempo of selection revealed in viral gene genealogies. BMC Evol. Biol. 11, 220 (2011).
Lemey, P., Rambaut, A., Drummond, A. J. & Suchard, M. A. Bayesian phylogeography finds its roots. PLOS Comput. Biol. 5, e1000520 (2009).
Fox, J. P., Hall, C. E., Cooney, M. K. & Foy, H. M. Influenzavirus infections in Seattle families, 1975–1979. I. Study design, methods and the occurrence of infections by time and age. Am. J. Epidemiol. 116, 212–227 (1982).
Longini, I. M., Jr, Koopman, J. S., Monto, A. S. & Fox, J. P. Estimating household and community transmission parameters for influenza. Am. J. Epidemiol. 115, 736–751 (1982).
Viboud, C. et al. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science 312, 447–451 (2006).
Recker, M., Pybus, O. G., Nee, S. & Gupta, S. The generation of influenza outbreaks by a network of host immune responses against a limited set of antigenic types. Proc. Natl Acad. Sci. USA 104, 7711–7716 (2007).
Zinder, D., Bedford, T., Gupta, S. & Pascual, M. The roles of competition and mutation in shaping antigenic and genetic diversity in influenza. PLoS Pathog. 9, e1003104 (2013).
Nobusawa, E. & Sato, K. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 80, 3675–3678 (2006).
Neuzil, K. M. et al. Immunogenicity and reactogenicity of 1 versus 2 doses of trivalent inactivated influenza vaccine in vaccine-naive 5–8-year-old children. J. Infect. Dis. 194, 1032–1039 (2006).
Matrosovich, M. N. et al. Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology 196, 111–121 (1993).
Hensley, S. et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326, 734–736 (2009).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Shapiro, B., Rambaut, A. & Drummond, A. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23, 7–9 (2006).
Shapiro, B. et al. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 28, 879–887 (2011).
Pagel, M., Meade, A. & Barker, D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 (2004).
Lemey, P., Minin, V. N., Bielejec, F., Pond, S. L. K. & Suchard, M. A. A counting renaissance: combining stochastic mapping and empirical Bayes to quickly detect amino acid sites under positive selection. Bioinformatics 28, 3248–3256 (2012).
Edwards, C. J. et al. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr. Biol. 21, 1251–1258 (2011).
Kelly, H. A., Grant, K., Williams, S., Fielding, J. & Smith, D. Epidemiological characteristics of pandemic influenza H1N1 2009 and seasonal influenza infection. Med. J. Aust. 191, 146–149 (2009).
Khiabanian, H., Farrell, G., St George, K. & Rabadan, R. Differences in patient age distribution between influenza A subtypes. PLoS ONE 4, e6832 (2009).
Mossong, J. et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 5, e74 (2008).
Rohani, P., Zhong, X. & King, A. A. Contact network structure explains the changing epidemiology of pertussis. Science 330, 982–985 (2010).
Bedford, T., Rambaut, A. & Pascual, M. Canalization of the evolutionary trajectory of the human influenza virus. BMC Biol. 10, 38 (2012).
Gog, J. R. & Grenfell, B. T. Dynamics and selection of many-strain pathogens. Proc. Natl Acad. Sci. USA 99, 17209–17214 (2002).
Lin, J., Andreasen, V., Casagrandi, R. & Levin, A. S. Traveling waves in a model of influenza A drift. J. Theor. Biol. 222, 437–445 (2003).
Smith, D. J. et al. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004).
Acknowledgements
We thank National Influenza Centres worldwide for their contributions to influenza virus surveillance. T.B. was supported by a Newton International Fellowship from the Royal Society and through National Institutes of Health (NIH) U54 GM111274. S.R. was supported by Medical Research Council (UK, Project MR/J008761/1), Wellcome Trust (UK, Project 093488/Z/10/Z), Fogarty International Centre (USA, R01 TW008246-01), Department of Homeland Security (USA, RAPIDD program), National Institute of General Medical Sciences (USA, MIDAS U01 GM110721-01) and National Institute for Health Research (UK, Health Protection Research Unit funding). The Melbourne WHO Collaborating Centre for Reference and Research on Influenza was supported by the Australian Government Department of Health and thanks N. Komadina and Y.-M. Deng. The Atlanta WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza was supported by the US Department of Health and Human Services. NIV thanks A.C. Mishra, M. Chawla-Sarkar, A. M. Abraham, D. Biswas, S. Shrikhande, B. AnuKumar, and A. Jain. Influenza surveillance in India was expanded, in part, through US Cooperative Agreements (5U50C1024407 and U51IP000333) and by the Indian Council of Medical Research. M.A.S. was supported through National Science Foundation DMS 1264153 and NIH R01 AI 107034. Work of the WHO Collaborating Centre for Reference and Research on Influenza at the MRC National Institute for Medical Research was supported by U117512723. P.L., A.R. & M.A.S were supported by EU Seventh Framework Programme [FP7/2007-2013] under Grant Agreement no. 278433-PREDEMICS and ERC Grant agreement no. 260864. C.A.R. was supported by a University Research Fellowship from the Royal Society.
Author information
Authors and Affiliations
Contributions
C.A.R. and T.B. conceived the research. C.A.R. and T.B. drafted the manuscript with substantial support from P.L. and S.R. I.G.B., S.B., M.C., N.J.C., R.S.D., C.P.G., A.C.H., A.K., A.Kl. X.L., J.W.M., T.O., V.P., Y.S., M.T., D.W. and X.X. coordinated and produced the influenza surveillance data. T.B. performed the modeling and data analyses along with C.A.R., S.R., P.L., M.A.S. and A.R. T.B. created the figures. All authors discussed the results and contributed to the revision of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
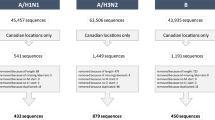
Extended Data Figure 1 Spatial distribution of 4,006 H3N2, 2,144 H1N1, 1,999 Vic and 1,455 Yam samples.
Circle area is proportional to the number of sequenced viruses originating from a location. Colour indicates assignment to one of 9 geographic regions.
Extended Data Figure 2 Inferred location of the trunk of H3N2 tree through time in the primary data set (a) and in a smaller secondary data set (b).
Coloured width at each time point indicates the posterior support for viruses from a particular geographic location comprising the trunk of the phylogenetic tree. Colours correspond to coloured circles in persistence insets in Fig. 1. The secondary data sets consist of 1,391 H3N2 viruses, 1,372 H1N1 viruses, 1,394 Vic viruses and 1,240 Yam viruses.
Extended Data Figure 3 Average inferred geographic history of region-specific samples for H3N2, former seasonal H1N1, Vic and Yam viruses from 2000 to 2012.
In each panel, phylogeny tips belonging to a particular region were collected and their phylogeographic histories traced backwards in time averaging across the phylogenetic tree to combine all viruses within each region. The x-axis shows number of years backward in time from phylogeny tips from a particular region and the y-axis shows the geographic make up as stacked histogram of the ancestors of these tips, where region colour-coding corresponds to the legend in Fig. 1. For example, the top left panel shows the ancestry of USA and Canadian H3N2 viruses. At x = 0, all of these viruses are still in the USA or Canada and so an unbroken yellow band takes up the entire y. However, at x = 1 year, a number of different geographic regions appear on the y. This indicates that, 1 year back, ancestors of USA and Canadian viruses are primarily found in Southeast Asia, India and South China. The pattern in the top right panel shows that the ancestors of USA and Canadian Yam viruses more often remain in the USA or Canada with approximately 50% of ancestors remaining 1 year back. Each panel is constructed by averaging across region-specific tips within a tree, but also across sampled posterior trees.
Extended Data Figure 4 Maximum clade credibility (MCC) trees for region-specific samples from USA/Canada, India and South China for H3N2, H1N1, Vic and Yam viruses.
Each tree only contains viruses from a particular geographic region and thus tips are all a single colour within a tree. Branch and trunk colouring have been retained from Fig. 1 to highlight the inferred geographic ancestry of each lineage.
Extended Data Figure 5 Antigenic map of Vic viruses primarily collected in 2008 (a), age distribution of infections for H3N2 (b), H1N1 (c) and B (d) in Australia 2000–2011, age distribution of ∼102.5 million passengers at London Heathrow and London Gatwick airports during 2011 (e), time series of virological characterizations from 2000 to 2012 of viruses from the USA by US CDC and from Australia by VIDRL for H3N2 (f), H1N1 (g), Vic (h) and Yam (i).
In a, the positions of strains (coloured circles) and antisera (uncoloured squares) are fit such that the distances between strains and antisera in the map represent the corresponding haemagglutination inhibition (HI) measurements with the least error following Smith et al.38 using data on Vic viruses from the WHO Collaborating Centre for Reference and Research on Influenza at the Centers for Disease Control and Prevention, Atlanta, Georgia, USA. Strains are coloured by antigenic cluster. Genetic clades corresponding to each antigenic cluster are marked with coloured vertical bars in Fig. 1c. The spacing between grid lines is one unit of antigenic distance corresponding to a twofold dilution of antiserum in the HI assay. In f–i, virological characterizations are a surrogate for epidemiological activity that allow for accurate discrimination among H3N2, H1N1, Vic, and Yam viruses. These data generally reflect the relative magnitudes and frequencies of epidemics but in some cases will inflate magnitudes of very small epidemics due to preferential characterization of subtypes circulating at low levels.
Extended Data Figure 6 Combined persistence estimates across pairs of regions for H3N2, H1N1, Vic and Yam (a) and Spearman correlation of a region’s persistence vs the region’s contribution to phylogenetic ancestry for H3N2, H1N1, Vic and Yam (b).
In a and b, persistence is measured as the average waiting time in years for a sample to leave its origin backwards in time in the phylogeny, with waiting time averaged across tips within a tree and across sampled posterior trees. In each panel of a, the diagonal shows persistence within each of the 9 study regions and within the combined region of ‘China’, for which nodes in North China and in South China were considered to belong to a single region. The estimates along the diagonal are equivalent to the means shown in Fig. 1. Off-diagonal elements show persistence estimates for pairwise combinations of regions. For example, the off-diagonal for North and South China is exactly equivalent to the diagonal element for ‘China’ and the off diagonal for ‘China’ and India represents mean persistence when combining nodes from North China, South China and India. In b, origin proportion is measured as the proportion of the time that a region is represented when tracing back 2 or more years from each tip in the phylogeny, averaged across tips within a tree and across sampled posterior trees. Spearman’s ρ is not significant for any individual virus. However, the probability of observing 4 instances where each virus has a ρ of at least 0.32 is significant (P = 0.0017, bootstrap resampling test).
Extended Data Figure 7 Simulation results for a model parameterized for slow antigenic drift (a), moderate antigenic drift (b), and fast antigenic drift (c).
Colours represent geographic regions with tropics in blue, north in yellow and south in red. Region-specific incidence patterns are shown in terms of cases per 100,000 individuals per week, patterns of antigenic drift in terms of increasing antigenic distance (roughly proportional to log2 HI units) over time and in the geographically labelled phylogeny. The parameterized antigenic mutation rate is 0.00015 antigenic mutations per infection per day in a, 0.00035 in b and 0.00055 in c, while the realized antigenic drift rate is 0.29 antigenic units per year in a, 0.58 in b and 1.19 in c. Between-region mixing is 5.26× faster in adults. Each panel shows output from a single simulation selected from the 112 shown in Fig. 3, and is intended to show model behaviours over a range of parameters, not necessarily the behaviour of particular viruses.
Extended Data Figure 8 Simulation results showing relationship between antigenic drift and persistence as a function of seasonality (a) and simulation results showing the effects of modulating transmission rate β on model behaviour (b).
In a, the seasonal forcing parameter ε follows ε = 0.00 (no forcing), ε = 0.04, ε = 0.08 and ε = 0.12 (moderate seasonal forcing). Points represent outcomes from a model in which adults travel between regions at 5.26× the rate of children. Solid black lines represent linear fits to the data. With 4 seasonality scenarios, 7 mutation rates and 8 replicates, there are 224 individual simulations shown. Persistence is measured as the average time in years taken for a tip to leave its region of origin going backwards in time, up the tree. In b, transmission rate β in contacts per day is varied and compared to its effect on observed antigenic drift (in antigenic units per year), attack rate per year, proportion of childhood infections and migration rate between regions (in events per viral lineage per year). One antigenic unit is roughly equivalent to one log2 HI unit. Black points represent outcomes from a model in which children and adults travel between regions at equal rates. Red points represent outcomes from a model in which adults travel between regions at 5.26× the rate of children. Solid black and red lines represent LOESS fits to the data. With 2 travel scenarios, 7 transmission rates and 8 replicates, there are 112 individual simulations shown.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and additional references. (PDF 102 kb)
Supplementary Data
This file contains the genetic sequence accession numbers. (XLSX 655 kb)
Source data
Rights and permissions
About this article
Cite this article
Bedford, T., Riley, S., Barr, I. et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 523, 217–220 (2015). https://doi.org/10.1038/nature14460
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14460
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.