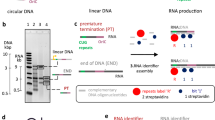
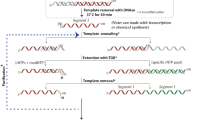
Abstract
Knowledge of the structure and dynamics of RNA molecules is critical to understanding their many biological functions. Furthermore, synthetic RNAs have applications as therapeutics and molecular sensors. Both research and technological applications of RNA would be dramatically enhanced by methods that enable incorporation of modified or labelled nucleotides into specifically designated positions or regions of RNA. However, the synthesis of tens of milligrams of such RNAs using existing methods has been impossible. Here we develop a hybrid solid–liquid phase transcription method and automated robotic platform for the synthesis of RNAs with position-selective labelling. We demonstrate its use by successfully preparing various isotope- or fluorescently labelled versions of the 71-nucleotide aptamer domain of an adenine riboswitch1 for nuclear magnetic resonance spectroscopy or single-molecule Förster resonance energy transfer, respectively. Those RNAs include molecules that were selectively isotope-labelled in specific loops, linkers, a helix, several discrete positions, or a single internal position, as well as RNA molecules that were fluorescently labelled in and near kissing loops. These selectively labelled RNAs have the same fold as those transcribed using conventional methods, but they greatly simplify the interpretation of NMR spectra. The single-position isotope- and fluorescently labelled RNA samples reveal multiple conformational states of the adenine riboswitch. Lastly, we describe a robotic platform and the operation that automates this technology. Our selective labelling method may be useful for studying RNA structure and dynamics and for making RNA sensors for a variety of applications including cell-biological studies, substance detection2, and disease diagnostics3,4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004)
Geiger, A., Burgstaller, P., von der Eltz, H., Roeder, A. & Famulok, M. RNA aptamers that bind l-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 24, 1029–1036 (1996)
Mairal, T. et al. Aptamers: molecular tools for analytical applications. Anal. Bioanal. Chem. 390, 989–1007 (2008)
Shangguan, D. et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 7, 2133–2139 (2008)
Wilson, C. & Keefe, A. D. Building oligonucleotide therapeutics using non-natural chemistries. Curr. Opin. Chem. Biol. 10, 607–614 (2006)
Usman, N., Ogilvie, K. K., Jiang, M. Y. & Cedergen, R. J. Automated chemical synthesis of long oligoribonucleotides using 2′-O-silylated ribonucleoside 3′-O-phosphoramidites on a controlled-pore glass support: synthesis of a 43-nucleotide sequence similar to the 3′-half molecule of an Escherichia coli formyl-methionine tRNA. J. Am. Chem. Soc. 109, 7845–7854 (1987)
Scaringe, S. A., Wincott, F. E. & Caruthers, M. H. Novel RNA synthesis method using 5′-O-silyl-2′-O-orthorster protection groups. J. Am. Chem. Soc. 120, 11820–11821 (1998)
Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 (1987)
Lu, K., Miyazaki, Y. & Summers, M. F. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR 46, 113–125 (2010)
Lyakhov, D. L. et al. Pausing and termination by bacteriophage T7 RNA polymerase. J. Mol. Biol. 280, 201–213 (1998)
Guo, Q., Nayak, D., Brieba, L. G. & Sousa, R. Major conformational changes during T7RNAP transcription initiation coincide with, and are required for, promoter release. J. Mol. Biol. 353, 256–270 (2005)
Sohn, Y., Shen, H. & Kang, C. Stepwise walking and cross-linking of RNA with elongating T7 RNA polymerase. Methods Enzymol. 371, 170–179 (2003)
Nudler, E., Gusarov, I. & Bar-Nahum, G. Methods of walking with the RNA polymerase. Methods Enzymol. 371, 160–169 (2003)
Pavlov, M. Y., Freistroffer, D. V. & Ehrenberg, M. Synthesis of region-labelled proteins for NMR studies by in vitro translation of column-coupled mRNAs. Biochimie 79, 415–422 (1997)
Marble, H. A. & Davis, R. H. RNA transcription from immobilized DNA templates. Biotechnol. Prog. 11, 393–396 (1995)
Huang, J., Brieba, L. G. & Sousa, R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry 39, 11571–11580 (2000)
Makarova, O. V., Makarov, E. M., Sousa, R. & Dreyfus, M. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl Acad. Sci. USA 92, 12250–12254 (1995)
Mentesana, P. E., Chin-Bow, S. T., Sousa, R. & McAllister, W. T. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J. Mol. Biol. 302, 1049–1062 (2000)
Leipply, D. & Draper, D. E. Dependence of RNA tertiary structural stability on Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry 49, 1843–1853 (2010)
Delfosse, V. et al. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 38, 2057–2068 (2010)
Noeske, J. et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA 102, 1372–1377 (2005)
Lee, M. K., Gal, M., Frydman, L. & Varani, G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl Acad. Sci. USA 107, 9192–9197 (2010)
Lemay, J. F., Penedo, J. C., Tremblay, R., Lilley, D. M. & Lafontaine, D. A. Folding of the adenine riboswitch. Chem. Biol. 13, 857–868 (2006)
Dalgarno, P. A. et al. Single-molecule chemical denaturation of riboswitches. Nucleic Acids Res. 41, 4253–4265 (2013)
Rieder, R., Lang, K., Graber, D. & Micura, R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem 8, 896–902 (2007)
Zhang, J. & Ferré-D'Amaré, A. R. Dramatic improvement of crystals of large RNAs by cation replacement and dehydration. Structure 22, 1363–1371 (2014)
Zhang, Q., Stelzer, A. C., Fisher, C. K. & Al-Hashimi, H. M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 450, 1263–1267 (2007)
Leipply, D. & Draper, D. E. Effects of Mg2+ on the free energy landscape for folding a purine riboswitch RNA. Biochemistry 50, 2790–2799 (2011)
Ferré-D'Amaré, A. R. & Doudna, J. A. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 24, 977–978 (1996)
Pinheiro, V. B. & Holliger, P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 32, 321–328 (2014)
Korbie, D. J. & Mattick, J. S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nature Protocols 3, 1452–1456 (2008)
Kao, C., Zheng, M. & Rüdisser, S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5, 1268–1272 (1999)
Mukherjee, S., Brieba, L. G. & Sousa, R. Structural transitions mediating transcription initiation by T7 RNA polymerase. Cell 110, 81–91 (2002)
Zuo, X. et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc. Natl Acad. Sci. USA 107, 1385–1390 (2010)
Ying, J. et al. Measurement of 1H-15N and 1H-13C residual dipolar couplings in nucleic acids from TROSY intensities. J. Biomol. NMR 51, 89–103 (2011)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Holmstrom, E. D. & Nesbitt, D. J. Single-molecule fluorescence resonance energy transfer studies of the human telomerase RNA pseudoknot: temperature-/urea-dependent folding kinetics and thermodynamics. J. Phys. Chem. B 118, 3853–3863 (2014)
Kapanidis, A. N. et al. Alternating-laser excitation of single molecules. Acc. Chem. Res. 38, 523–533 (2005)
Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008)
Cordes, T., Vogelsang, J. & Tinnefeld, P. On the mechanism of Trolox as antiblinking and antibleaching reagent. J. Am. Chem. Soc. 131, 5018–5019 (2009)
Fiore, J. L., Hodak, J. H., Piestert, O., Downey, C. D. & Nesbitt, D. J. Monovalent and divalent promoted GAAA tetraloop-receptor tertiary interactions from freely diffusing single-molecule studies. Biophys. J. 95, 3892–3905 (2008)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012)
Steinberg, G., Stromsborg, K., Thomas, L., Barker, D. & Zhao, C. Strategies for covalent attachment of DNA to beads. Biopolymers 73, 597–605 (2004)
Chivers, C. E. et al. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nature Methods 7, 391–393 (2010)
Acknowledgements
We thank D. E. Draper, A. Bax, A. Byrd, M. Summers, A. Rein, and J. Strathern for discussions. This work was supported in part by the Intramural Research Programs of the National Cancer Institute, the National Institute of Diabetes, Digestive and Kidney Diseases, the National Heart, Lung and Blood Institute; by the Intramural Antiviral Target Program (IATAP) of the Office of the Director, National Institutes of Health; by the 2013 Director’s Challenge Innovation Award of the National Institutes of Health; by the fund from the National Cancer Institute under contract number HHSN261200800001E; by the National Science Foundation (CHE1266416, PHYS1125844); by the National Institutes of Health Molecular Biophysics Training Program (T32 GM-065103); and by the W. M. Keck Foundation, and the National Institute of Standards and Technology. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Author information
Authors and Affiliations
Contributions
Y.L. and P.Y. performed RNA synthesis and NMR experiments; E.H. and D.J.N. designed and performed smFRET experiments; J.W. contributed to chemical shift assignments; J.Z. and A.R.F. helped to design the DNA template using PCR, crystallized and determined the three-dimensional structure of the PLOR-generated riboA71; J.Y., M.A.D., D.C., and S.L. helped to characterize RNA; R.S. provided critical advice about T7-enzymatic synthesis and revised the manuscript; J.R.S. helped to characterize RNAs and revised the manuscript; Y.L., E.H., J.Z., J.R.S., and Y.-X.W. prepared figures; Y.-X.W. designed PLOR and the automated platform and wrote the manuscript. All authors discussed the results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Synthesis algorithm for the selective labelling of RNA using the PLOR method.
All samples except for fully labelled 71-nt‐CN, S1+Lk1‐CN, S1+Lk1‐H, and 15N‐104-nt‐TCV were synthesized following this algorithm. The initiation, elongation, and termination stages are shown in green, blue, and red, respectively. Various NTP combinations added during the elongation cycles depend on the desired labelling scheme.
Extended Data Figure 2 Automated platform for PLOR synthesis.
a, Diagram of the automated platform for PLOR synthesis depicting its various parts and stations. b, Top‐view (left) and side‐view (right) photographs of the in‐house automated platform.
Extended Data Figure 3 Optimization of experimental conditions for PLOR synthesis.
a, The products of the PLOR (left) and the standard transcription methods (right). PLOR generates a pure full‐length product with the desired labelling in the final step. b, Comparison of PLOR efficiency for Lp2‐CN synthesis under various conditions: I, freshness of DNA‐attached beads; II, anion specificity; III, [Mg2+]; IV, K2SO4 presence in the initiation; V, increasing NTPs (‘1X’ represents NTP amounts in Extended Data Table 1); VI, ratio of ATP/GTP:DNA; VII, ratio of UTP:DNA; and VIII, incubation time of the termination. The right lane contains pure RiboA71 as a control.
Extended Data Figure 4 Comparison of NMR spectra of PLOR‐generated and 71-nt‐CN samples in adenine‐bound form.
Superposition of the 1H13C‐TROSY spectra of 71-nt‐CN with Lp1‐CN (a), Lp2‐CN (b), Lp1+2‐CN (c), Lk2‐CN (d), S1+Lk1‐CN (e), and 4nt‐CN (f). These results indicate that the RNAs synthesized by PLOR have the same fold as 71-nt‐CN generated using the traditional solution‐based transcription method and are functional as evidenced by binding of the adenine ligand.
Extended Data Figure 5 PLOR‐generated riboA71 maintains both sequence and structural fidelity.
a, Structural superposition of the PLOR‐generated riboA71 (PDB accession number 4XNR) with that of the riboA71 generated using the regular in vitro transcription (PDB accession number 4TZX)26. The root mean squared deviation between all C1 atoms was ~ 0.3 Å. b, Structural superposition of the PLOR‐generated riboA71 (PDB accession number 4XNR) with that of the riboA71 (PDB accession number 1Y26)1. The root mean squared deviation between all C1 atoms was ~ 0.4 Å. c, Sequences and secondary structures of the RNAs in b. The arrows denote nucleotide sequence differences between the two ribA71 sequences. d, Composite simulated anneal‐omit 2|Fo| − |Fc| electron density of the riboA71 RNA structure (PDB accession number 4XNR) at 2.2 Å resolution calculated using the final model (1.0 s.d.). e, Portion of the electron density in c in the adenine‐binding pocket unambiguously identifies the nucleobase identities of the PLOR‐generated RNA, revealing no undesired sequence changes introduced by PLOR. f, Portion of the electron density in c of the G6·C66 base pair, which differs in identity between the two structures, suggesting that if there had been undesired sequence substitutions that resulted from using the PLOR method, they would have been readily detected in the crystallographic analysis.
Extended Data Figure 6 Using PLOR to isotopically label a 104‐nt RNA.
a, Secondary structure of the 104‐nt structural element in the TCV genomic RNA. The 15N‐H1‐TCV was synthesized by the PLOR method, and the sequence in red is 15N‐labelled in 15N‐H1‐TCV. b, 15N‐TROSY spectrum of 15N‐H1‐TCV RNA. c, Superposition of the 1H15N‐TROSY spectra of 15N‐H1‐TCV with 15N‐104-nt‐TCV34, indicating that the PLOR‐generated selectively labelled RNA has the same fold as that generated using the regular in vitro transcription.
Rights and permissions
About this article
Cite this article
Liu, Y., Holmstrom, E., Zhang, J. et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature 522, 368–372 (2015). https://doi.org/10.1038/nature14352
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14352
This article is cited by
-
Extending the toolbox for RNA biology with SegModTeX: a polymerase-driven method for site-specific and segmental labeling of RNA
Nature Communications (2023)
-
Structural and dynamic mechanisms for coupled folding and tRNA recognition of a translational T-box riboswitch
Nature Communications (2023)
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Bioorthogonal information storage in l-DNA with a high-fidelity mirror-image Pfu DNA polymerase
Nature Biotechnology (2021)
-
Metal ions and sugar puckering balance single-molecule kinetic heterogeneity in RNA and DNA tertiary contacts
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.