Abstract
Many acute and chronic anaemias, including haemolysis, sepsis and genetic bone marrow failure diseases such as Diamond–Blackfan anaemia, are not treatable with erythropoietin (Epo), because the colony-forming unit erythroid progenitors (CFU-Es) that respond to Epo are either too few in number or are not sensitive enough to Epo to maintain sufficient red blood cell production1,2,3,4,5,6,7,8,9. Treatment of these anaemias requires a drug that acts at an earlier stage of red cell formation and enhances the formation of Epo-sensitive CFU-E progenitors. Recently, we showed that glucocorticoids specifically stimulate self-renewal of an early erythroid progenitor, burst-forming unit erythroid (BFU-E), and increase the production of terminally differentiated erythroid cells10,11. Here we show that activation of the peroxisome proliferator-activated receptor α (PPAR-α) by the PPAR-α agonists GW7647 and fenofibrate synergizes with the glucocorticoid receptor (GR) to promote BFU-E self-renewal. Over time these agonists greatly increase production of mature red blood cells in cultures of both mouse fetal liver BFU-Es and mobilized human adult CD34+ peripheral blood progenitors, with a new and effective culture system being used for the human cells that generates normal enucleated reticulocytes. Although Ppara−/− mice show no haematological difference from wild-type mice in both normal and phenylhydrazine (PHZ)-induced stress erythropoiesis, PPAR-α agonists facilitate recovery of wild-type but not Ppara−/− mice from PHZ-induced acute haemolytic anaemia. We also show that PPAR-α alleviates anaemia in a mouse model of chronic anaemia. Finally, both in control and corticosteroid-treated BFU-E cells, PPAR-α co-occupies many chromatin sites with GR; when activated by PPAR-α agonists, additional PPAR-α is recruited to GR-adjacent sites and presumably facilitates GR-dependent BFU-E self-renewal. Our discovery of the role of PPAR-α agonists in stimulating self-renewal of early erythroid progenitor cells suggests that the clinically tested PPAR-α agonists we used may improve the efficacy of corticosteroids in treating Epo-resistant anaemias.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kaushansky K. et al. Williams Hematology 8th edn (McGraw Hill, 2010)
Livingston, D. H. et al. Bone marrow failure following severe injury in humans. Ann. Surg. 238, 748–753 (2003)
Jones, K. B., Anderson, D. W. & Longmore, G. D. Effects of recombinant hematopoietins on blood-loss anemia in mice. Iowa Orthop. J. 25, 129–134 (2005)
Robinson, Y. et al. Impaired erythropoiesis after haemorrhagic shock in mice is associated with erythroid progenitor apoptosis in vivo . Acta Anaesthesiol. Scand. 52, 605–613 (2008)
Zimmerman, J. L. Use of blood products in sepsis: an evidence-based review. Crit. are Med. 32, S542–S547 (2004)
Fiorillo, A. et al. Unresponsiveness to erythropoietin therapy in a case of Blackfan Diamond anemia. Am. J. Hematol. 37, 65 (1991)
Nathan, D. G., Clarke, B. J., Hillman, D. G., Alter, B. P. & Housman, D. E. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. J. Clin. Invest. 61, 489–498 (1978)
Vlachos, A. & Muir, E. How I treat Diamond-Blackfan anemia. Blood 116, 3715–3723 (2010)
Niemeyer, C. M. et al. Treatment trial with recombinant human erythropoietin in children with congenital hypoplastic anemia. Contrib. Nephrol. 88, 276–280 (1991)
Flygare, J., Rayon Estrada, V., Shin, C., Gupta, S. & Lodish, H. F. HIF1α synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 117, 3435–3444 (2011)
Zhang, L. et al. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature 499, 92–96 (2013)
Willig, T. N. et al. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. Pediatr. Res. 46, 553–561 (1999)
Lipton, J. M., Atsidaftos, E., Zyskind, I. & Vlachos, A. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr. Blood Cancer 46, 558–564 (2006)
Bauer, A. et al. The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev. 13, 2996–3002 (1999)
von Lindern, M. et al. The glucocorticoid receptor cooperates with the erythropoietin receptor and c-Kit to enhance and sustain proliferation of erythroid progenitors in vitro . Blood 94, 550–559 (1999)
O’Malley, B. The steroid receptor superfamily: more excitement predicted for the future. Mol. Endocrinol. 4, 363–369 (1990)
Moore, J. T., Collins, J. L. & Pearce, K. H. The nuclear receptor superfamily and drug discovery. ChemMedChem 1, 504–523 (2006)
Yang, L. P. & Keating, G. M. Fenofibric acid: in combination therapy in the treatment of mixed dyslipidemia. Am. J. Cardiovasc. Drugs 9, 401–409 (2009)
Muoio, D. M. et al. Peroxisome proliferator-activated receptor-α regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes 51, 901–909 (2002)
Lee, S. S. et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15, 3012–3022 (1995)
Flygare, J. et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood 105, 4627–4634 (2005)
Ebert, B. L. et al. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood 105, 4620–4626 (2005)
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013)
Wong, P. et al. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood 118, e128–e138 (2011)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010)
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010)
Acknowledgements
We thank the Whitehead Institute Flow Cytometry Facility, Genome Technology Core and Bioinformatics & Research Computing Facility, as well as the Massachusetts Institute of Technology Koch Institute Flow Cytometry Core. We thank V. Sankaran for haemoglobin HPLC and J. Flygare for the plasmid encoding the RPS19 shRNA, and are grateful to animal technicians F. Reinhardt and T. E. Chavarria for their assistance. We thank T. DiCesare for assistance with graphics. This study was supported by grants to H.F.L. (Defense Advanced Research Projects Agency HR0011-14-2-0005; Department of Defense/US Army Medical Research and Materiel Command W81WH-12-1-0449, National institutes of Health (NIH)/National Heart, Lung, and Blood Institute 2 P01 HL032262-25; as well as research support from the Diamond-Blackfan Anemia Foundation and Diamond Blackfan Anemia Canada. L.L.P. was supported by NIH grant DK100692. X.G. was supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Contributions
H.-Y.L., X.G., L.L.P. and H.F.L. designed the experiments. H.-Y.L., X.G. and R.R.E. performed the experiments. M.I.B. and H.L. conducted bioinformatic analyses of ChIP-seq and RNA-seq. H.-Y.L., X.G. and H.F.L. wrote the manuscript with input from M.I.B. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 PPAR-α agonist GW7647 does not have adverse effects on erythroid differentiation and has no effects on CFU-E cells.
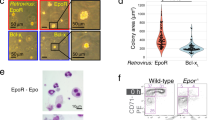
a, Gene expression changes of nuclear receptors in BFU-E cells from RNA-seq results published previously10. b, Flow cytometry analyses of CD71 and Ter119 markers to demonstrate erythroid differentiation of mouse BFU-E cells after 9 days of culture with the indicated additions. c, Ppara gene expression in BFU-E, CFU-E and Ter119+ erythroblasts. BFU-E, CFU-E and Ter119+ erythroid cells were isolated from E14.5 mouse fetal livers as described24. Total RNA was purified for quantitative PCR analysis. Ppara gene expression was normalized to mouse 18S rRNA in different stages. Error bars represent mean ± s.d. from three independent experiments. d, DNase I hypersensitivity (HS) analysis at Ppara promoter region in different mouse cells from Encode. e, Production of mouse erythroblasts from isolated CFU-E cells. Wild-type mouse CFU-E cells from E14.5 fetal livers were untreated (black line) or treated with DEX (blue line), GW7647 (red line) or fenofibrate (green line). Error bars represent mean ± s.d. from three independent experiments. f, Colony-forming assays were conducted at 48 h after compound treatment to determine BFU-E colony numbers from 100 mouse BFU-E cells cultured under the indicated conditions. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. g, At day 3, BFU-E colony numbers from 100 purified mouse BFU-E cells were quantified by colony forming assays. 100 purified mouse BFU-E cells were untreated or treated with DEX alone or DEX in combination with agonists or antagonists targeting PPAR receptors (α, γ or β). BFU-E colonies were quantified after 8 days in culture. *P < 0.05, **P < 0.01, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. h, Real-time PCR analysis of gene expression in DEX-treated and DEX+GW7647-treated wild-type or Ppara−/− mouse BFU-E cells. * P < 0.05, *** P < 0.001, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments.
Extended Data Figure 2 Human CD34+ erythroid differentiation system.
a, Total CFU-E colonies formed during days 0–9. CFU-E colony numbers were quantified by plating 1,000 cells from various time points during days 0–9 of the human CD34+ erythroid culture on methylcellulose. CFU-E colonies were quantified after 12–14 days. Total CFU-E colony numbers in culture under conditions without GW7647 (black line) or with GW7647 (red line) were calculated using the total cell numbers at corresponding time points in Fig. 2a. b, Human CD34+ cells were treated at day 1 with 100 nM GW7647 with or without DEX at the concentration indicated in the figure. At day 6, total cell numbers were counted and cells were collected for BFU-E colony assays. c, Protein expression of PPAR-α demonstrating shRNA knockdown efficiency via lentiviral transduction. LacZ shRNA is used as a control. shRNA-1 and -2 are both specific for PPAR-α. shRNA-2 has higher knockdown efficiency. d, Cell pellets of 1 million cells demonstrating haemoglobin accumulation during the differentiation process. e, Flow cytometry analyses of erythroid markers during the 21-day human CD34+ erythroid culture. Top, c-KIT versus CD235a; middle, CD71 versus CD235a. Note the sequential induction of c-kit, CD71 and CD235a, as well as the sequential downregulation of c-kit and CD71. Bottom, enucleated reticulocytes are CD235a+Hoechst−, nuclei are CD235a−Hoechst+, and nucleated erythroblasts are CD235a+Hoechst+. Enucleation rate is 32.6/(32.6+37.7) × 100% = 46.4%. f, Summary of high-performance liquid chromatography (HPLC) results using haemolysates of cultured reticulocytes and normal human RBCs (control). Top, total protein composition of haemolysates. Bottom, haemoglobin composition of haemolysates. Cultured reticulocytes contain more than 90% of adult globins. g, Size measurement of enucleated reticulocytes by both diameter and area. Scale bar, 10 µm. h, Benzidine–Giemsa staining of human reticulocytes cultured with or without GW7647. Scale bar, 12 µm. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments.
Extended Data Figure 3 GW7647 increases erythroid progenitors and CD235a+ cells in RPS19-knockdown human progenitor cells.
a, Human CD34+ haematopoietic progenitors were transduced with lentivirus encoding GFP and either a scrambled shRNA or an shRNA targeting RPS19. Then transduced cells were treated with or without 100 nM GW7647. After 48 h, GFP+ cells were sorted by FACS and plated for BFU-E and CFU-E colony-forming assays. RPS19 knockdown efficiency is shown at the bottom. M.W., molecular weight. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. b, Sorted GFP+ cells were returned to culture with the indicated concentration of GW7647. Top, percentage of CD71+ cells at day 9 was determined by FACS. Bottom, percentage of CD235a+ cells at day 21 was determined by FACS. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. c. Total cell numbers generated from one GFP-positive cell at the indicated times of culture. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments.
Extended Data Figure 4 GW7647 improves anaemia in two mouse models of anaemia.
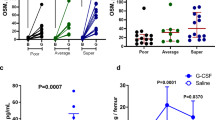
a, Experimental scheme for PHZ- induced haemolytic anaemia, used also in Fig. 3a. Wild-type or Ppara−/− mice were pretreated with DMSO (control) or GW7647 (100 μg kg−1) for 3 days (days −3 to −1) before PHZ injection on day 0. Mice were subject to daily DMSO or GW7647 injections during days 0–6. Red arrows indicate days of blood sample collection. b, Ppara−/− mice were treated with DMSO or GW7647 and then injected with PHZ. Haemoglobin (HGB), red blood cell numbers (RBC) and haematocrit (HCT) were measured on the days indicated. Error bars represent mean ± s.d. from six mice. c, BFU-E and CFU-E colony-forming assays of spleen or bone marrow cells. Wild-type or Ppara−/− mice were treated with PHZ and DMSO (control) or GW7647 (100 μg kg−1) as described in earlier. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. d, Spleen and bone marrow cells were harvested. Representative flow cytometry analysis of spleen erythroblasts isolated from GW7647- or DMSO-treated wild-type mice at day 3 after PHZ injection. FSC-A, forward scatter area.
Extended Data Figure 5 GW7647 increases BFU-E numbers in Nan/+ mutant mice.
a, Corticosteroid levels in serum were measured in wild-type and Nan/+ mutant mice. Each dot represents one mouse. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from all mice. b, Increase of BFU-E numbers in spleens from GW7647-treated wild-type and Nan/+ mutant mice at day 18. *P < 0.05, Student’s t-test. c, Total numbers of white blood cells (WBC) and platelets from peripheral blood samples were measured at day 0 and day 18. Each dot represents one mouse. Error bars represent mean ± s.d. from all mice.
Extended Data Figure 6 Bioinformatic analyses of mouse BFU-E cells.
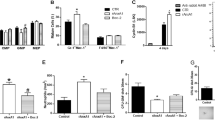
a, Genome-wide distribution of GR and PPAR-α chromatin occupancy sites in BFU-E cells. ChIP-seq analyses of GR and PPAR-α occupancy in mouse BFU-E cells isolated from DEX- and GW7647-treated wild-type E14.5 fetal livers. TSS, transcription start site; TTS, transcription termination site; UTR, untranslated region. Distal intergenic, regions greater than 3 kb from TSS. b, Venn diagram showing overlap between GR and PPAR-α chromatin occupancy sites. c, De novo motif searching of the overlapped chromatin sites occupied by GR and PPAR-α. The table depicts transcription-factor-binding motifs enriched at GR and PPAR-α overlapping sites relative to genomic background and associated P values. d, Real-time PCR analysis of Pu.1 gene expression in mouse BFU-E cells transduced with virus encoding either LacZ shRNA or Pu.1 shRNA. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. e, Colony-forming assays were conducted to determine BFU-E colony numbers from 100 mouse BFU-E cells infected with virus encoding either LacZ shRNA or Pu.1 shRNA. Cells were cultured in SFELE medium with or without DEX ± GW7647. Colony-forming assays were performed at 48 h. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments.
Extended Data Figure 7 Kit is a target gene of GR and PPAR-α.
a, Real-time RT–PCR analysis of Kit gene expression in wild-type and Ppara−/− mouse BFU-E cells untreated or treated with DEX with or without the addition of GW7647. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. b, Human CD34+ cells were treated with or without GW7647 as described in Fig. 2. Top, at day 9 of culture, cell-surface KIT and CD71 expression were analysed by flow cytometry. Bottom, a representative histogram of KIT expression in cells treated or untreated with GW7647. c, ChIP-seq occupancy signal map of GR and PPAR-α across the Kit locus in BFU-E cells.
Extended Data Figure 8 Pathways modulated by PPAR-α activation.
Pathway analysis of RNA-seq data of genes that are up- or downregulated by more than 50%, comparing cultures treated with DEX alone or DEX+GW7647.
Extended Data Figure 9 PPAR-α antagonist interferes with the function of PPAR-α but not GR.
a, Quantitative ChIP analysis of GR and PPAR-α occupancy at Kit and Ppara loci in mouse BFU-E cells following the indicated treatments. Units are arbitrary; signals using rabbit IgG are represented by grey dot lines across the plots. *P < 0.05, **P < 0.01, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments. b, Co-immunoprecipitation (IP) measuring interaction between GR and PPAR-α in mouse BFU-E cells isolated from E14.5 fetal livers in wild-type or Ppara−/−mice. BFU-E cells were untreated, or treated with DEX with or without GW7647 with or without GW6471. Whole-cell lysates were incubated with anti-GR antibody and immunoprecipitates were probed with specific antibodies as indicated. M.W., molecular weight. c, Colony-forming assays to determine BFU-E colony numbers from 100 mouse BFU-E cells cultured with the indicated treatments. *P < 0.05, Student’s t-test. Error bars represent mean ± s.d. from three independent experiments.
Extended Data Figure 10 Model of synergism between PPAR-α and GR to promote BFU-E self-renewal.
BFU-E cells normally undergo limited self-renewal to generate CFU-Es, which can differentiate into erythroblasts. GR is sequestered in the cytoplasm without glucocorticoids such as DEX. Upon glucocorticoid treatment, liganded GR will be translocated into the nucleus and bind to chromatin to regulate gene transcription important for BFU-E self-renewal. PPAR-α is often recruited to chromatin sites that are in close proximity to GR by glucocorticoid treatment alone without any function on BFU-E self-renewal. Upon glucocorticoid and PPAR-α agonist co-treatment, activated PPAR-α interacts with GR to modulate GR transcriptional activity. This leads to enhanced BFU-E self-renewal, and over time generates more CFU-Es and erythroblasts.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and Supplementary References. (PDF 325 kb)
Supplementary Table 1
This table contains RNA-Seq analysis from previous studies (ref. 10). Genes with gene expression changes above or below 2 fold are included here. Gene ID, symbols and description are also included. (XLSX 235 kb)
Supplementary Table 2
This table shows gene expression changes in mouse BFU-Es treated with GW7647 and DEX versus DEX alone. Mouse BFU-Es were isolated from E14.5 embryos. BFU-Es were treated with or without DEX plus or minus GW7647 for 12 hrs. Cells were harvested and RNA were extracted for RNA-Seq analysis. (XLSX 43 kb)
Supplementary Table 3
This table contains complete blood count analysis of wild-type and PPARα-/- mice. (XLSX 37 kb)
Supplementary Table 4
This table shows gene expression changes in mouse BFU-Es treated with GW7647 and DEX versus DEX alone. Mouse BFU-Es were isolated from E14.5 embryos. BFU-Es were treated with or without DEX plus or minus GW7647 for 12 hrs. Cells were harvested and RNA were extracted for RNA-Seq analysis. (XLSX 991 kb)
Supplementary table 5
This table contains lists of up- or down-regulated genes (DEX and GW7647 versus DEX) with 10 kb of GR and PPARa binding peaks. (XLSX 13 kb)
Supplementary Table 6
This table contains raw data for Figure 3a and Extended Data Figure 4b. (XLSX 44 kb)
Supplementary Table 7
This table contains raw data for Figure 3b and Extended Data Figure 5c. (XLSX 38 kb)
Rights and permissions
About this article
Cite this article
Lee, HY., Gao, X., Barrasa, M. et al. PPAR-α and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature 522, 474–477 (2015). https://doi.org/10.1038/nature14326
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14326
This article is cited by
-
Nucleated red blood cells explain most of the association between DNA methylation and gestational age
Communications Biology (2023)
-
Heterochromatin rewiring and domain disruption-mediated chromatin compaction during erythropoiesis
Nature Structural & Molecular Biology (2023)
-
ARID1B, a molecular suppressor of erythropoiesis, is essential for the prevention of Monge’s disease
Experimental & Molecular Medicine (2022)
-
Interactions between nuclear receptors glucocorticoid receptor α and peroxisome proliferator–activated receptor α form a negative feedback loop
Reviews in Endocrine and Metabolic Disorders (2022)
-
Proteomic analysis provides insights into the function of Polian vesicles in the sea cucumber Apostichopus japonicus post-evisceration
Journal of Oceanology and Limnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.