Abstract
Disruption of the MECP2 gene leads to Rett syndrome (RTT), a severe neurological disorder with features of autism1. MECP2 encodes a methyl-DNA-binding protein2 that has been proposed to function as a transcriptional repressor, but despite numerous mouse studies examining neuronal gene expression in Mecp2 mutants, no clear model has emerged for how MeCP2 protein regulates transcription3,4,5,6,7,8,9. Here we identify a genome-wide length-dependent increase in gene expression in MeCP2 mutant mouse models and human RTT brains. We present evidence that MeCP2 represses gene expression by binding to methylated CA sites within long genes, and that in neurons lacking MeCP2, decreasing the expression of long genes attenuates RTT-associated cellular deficits. In addition, we find that long genes as a population are enriched for neuronal functions and selectively expressed in the brain. These findings suggest that mutations in MeCP2 may cause neurological dysfunction by specifically disrupting long gene expression in the brain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chahrour, M. & Zoghbi, H. Y. The story of Rett syndrome: from clinic to neurobiology. Neuron 56, 422–437 (2007)
Guy, J., Cheval, H., Selfridge, J. & Bird, A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 27, 631–652 (2011)
Tudor, M., Akbarian, S., Chen, R. Z. & Jaenisch, R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc. Natl Acad. Sci. USA 99, 15536–15541 (2002)
Jordan, C., Li, H. H., Kwan, H. C. & Francke, U. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med. Genet. 8, 36 (2007)
Chahrour, M. et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229 (2008)
Ben-Shachar, S., Chahrour, M., Thaller, C., Shaw, C. A. & Zoghbi, H. Y. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum. Mol. Genet. 18, 2431–2442 (2009)
Samaco, R. C. et al. Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nature Genet. 44, 206–211 (2012)
Baker, S. A. et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell 152, 984–996 (2013)
Zhao, Y. T., Goffin, D., Johnson, B. S. & Zhou, Z. Loss of MeCP2 function is associated with distinct gene expression changes in the striatum. Neurobiol. Dis. 59, 257–266 (2013)
King, I. F. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 (2013)
Sugino, K. et al. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci. 34, 12877–12883 (2014)
Meins, M. et al. Submicroscopic duplication in Xq28 causes increased expression of the MECP2 gene in a boy with severe mental retardation and features of Rett syndrome. J. Med. Genet. 42, e12 (2005)
Collins, A. L. et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum. Mol. Genet. 13, 2679–2689 (2004)
Lyst, M. J. et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nature Neurosci. 16, 898–902 (2013)
Li, Y. et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell 13, 446–458 (2013)
Deng, V. et al. FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum. Mol. Genet. 16, 640–650 (2007)
Lewis, J. D. et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69, 905–914 (1992)
Mellén, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012)
Guo, J. U. et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nature Neurosci. 17, 215–222 (2014)
Skene, P. J. et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 (2010)
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009)
Szulwach, K. E. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature Neurosci. 14, 1607–1616 (2011)
Xie, W. et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 (2012)
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013)
Cohen, S. et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72, 72–85 (2011)
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004)
Polymenidou, M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature Neurosci. 14, 459–468 (2011)
Raychaudhuri, S. et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 6, e1001097 (2010)
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011)
Yazdani, M. et al. Disease modeling using embryonic stem cells: MeCP2 regulates nuclear size and RNA synthesis in neurons. Stem Cells 30, 2128–2139 (2012)
McGill, R., Tukey, J. & Larsen, W. A. Variations of box plots. Am. Stat. 32, 12–16 (1978)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Kim, T. K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578 (2012)
Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in Biosciences 131, 281–285 (2012)
Wu, Y., Genton, M. G. & Stefanski, L. A. A multivariate two-sample mean test for small sample size and missing data. Biometrics 62, 877–885 (2006)
Chen, P. Y., Cokus, S. J. & Pellegrini, M. B. S. Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics 11, 203 (2010)
Ebert, D. H. et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499, 341–345 (2013)
Feng, J., Liu, T. & Zhang, Y. Using MACS to identify peaks from ChIP-seq data. Current Protocols in Bioinformat. Chapter 2, Unit 2 14. (2011)
Nagy, G., Daniel, B., Jonas, D., Nagy, L. & Barta, E. A novel method to predict regulatory regions based on histone mark landscapes in macrophages. Immunobiology 218, 1416–1427 (2013)
Tronche, F. et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nature Genet. 23, 99–103 (1999)
Chen, W. G. et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889 (2003)
Huang da. W, Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Brown, V. et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001)
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods 5, 621–628 (2008)
Gray, J. M. et al. SnapShot-Seq: a method for extracting genome-wide, in vivo mRNA dynamics from a single total RNA sample. PLoS ONE 9, e89673 (2014)
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nature Protocols 1, 241–245 (2006)
Zhou, Z. et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269 (2006)
Valinluck, V. et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 32, 4100–4108 (2004)
Hashimoto, H. et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849 (2012)
Spruijt, C. G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013)
Khrapunov, S. et al. Unusual characteristics of the DNA binding domain of epigenetic regulatory protein MeCP2 determine its binding specificity. Biochemistry 53, 3379–3391 (2014)
Acknowledgements
We thank E. Griffith and members of the Greenberg laboratory, A. Bird, G. Mandel and members of their laboratories, and M. Coenraads for discussions, M. Goodell for providing the Dnmt3a mice, N. Sharma and F. DiBiase for experimental support, and M. Mistry of the HSPH Bioinformatics Core, Harvard School of Public Health for assistance with gene expression analysis. This work was supported by grants from the Rett Syndrome Research Trust and the National Institutes of Health (NIH) (1RO1NS048276) to M.E.G., fellowships from the Damon Runyon Cancer Research Foundation (DRG-2048-10) and the William Randolf Hearst fund to H.W.G., as well as NIH grant T32GM007753, and the HHMI Gilliam fellowship to B.K., H.S. is a HHMI Fellow of the Damon Runyon Cancer Research Foundation (DRG-2194-14).
Author information
Authors and Affiliations
Contributions
H.W.G. and B.K. performed or directed all experiments and analysis in the study. H.W.G., B.K. and D.A.H. performed gene expression analysis. B.K. performed EMSA assays. H.W.G. performed ChIP-seq analysis. H.W.G., H.S., N.R.K. performed bisulfite sequencing and DNA methylation analysis. H.W.G., D.A.H., H.S., N.R.K. and M.H. performed bioinformatics and statistical analysis. H.W.G., B.K. and C.S.G. performed Dnmt3a mouse experiments and neuronal culture experiments. D.H.E. provided mouse reagents. H.W.G., B.K. and M.E.G. wrote the manuscript. M.E.G. advised on all aspects of the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Analysis of gene expression changes in Mecp2 mutant mice.
a, Heatmap of median gene lengths for genes identified as misregulated in Mecp2 mutant studies or sixteen different studies of neurological dysfunction and disease in mice. Mouse model and GEO accession number, or reference, are listed (for Strand et al. (1), 3NP treatment; (2), human HD brain; (3), R2/6 Htt transgenic). b, Scatter plots of fold-change in gene expression in the MeCP2 KO for the amygdala (left), which shows robust length-dependent misregulation, and the liver (right), which does not. Fold-change values for genes (black points) and mean fold-change for 200 genes per bin with a 40 gene step are shown (mean, red line; ribbon, s.e.m.). c, The fraction of genes showing fold-change >0 for data sets in b; genes binned by length (100 gene bins, 50 gene step). d–f, Analysis of published microarray5,6,7,8,9 (d, e) or RNA sequencing (RNA-seq)18 (f) data sets from MeCP2 KO (d, f) or OE (e) mice. Mean fold-change in expression (200 gene bins, 40 gene step), red line; ribbon, s.e.m. For d–f, mean (black line) and two standard deviations (grey ribbon) are shown for 10,000 resamplings in which gene lengths were randomized with respect to fold-change. The spike in mean fold-change at ∼1 kb in several plots corresponds to the olfactory receptor genes (Supplementary Discussion). g, Mean changes in expression of genes binned by length from RNA-seq analysis of MeCP2 KO cortex (n = 3 per genotype). h, Mean changes in expression from microarray analysis of genes binned by length in MeCP2(R306C) cerebellum (n = 4 per genotype) i, Heatmap summary of fold-changes in gene expression from RNA-seq analysis of Mecp2 mutant mean in g compared to Nanostring nCounter (18 genes, top) or RT-qPCR (17 genes, bottom) analysis from cortex (n = 4 per genotype). Selected long genes (>100 kb) consistently upregulated in the MeCP2 KO or downregulated in MeCP2 OE mutant mice across brain tissues were tested (Supplementary Table 2). A statistically significant upregulation of these genes is observed in the cortex for both MeCP2 KO (nCounter, P = 0.00073; qPCR, P < 1 × 10−15) and MeCP2(R306C) (nCounter, P = 0.0482 ; qPCR, P = 1.69 × 10−6; Hotelling T2 test for small sample size37). Note that for completeness, data from other figures have been re-presented here. See Methods and Supplementary Table 1 for sample sizes from published data sets and other details.
Extended Data Figure 2 Timing and severity of gene expression changes in models of RTT.
a, Mean fold-change in gene expression versus gene length in the hippocampus of MeCP2 KO mice compared to wild type at four and nine weeks of age reveals increasing magnitude of length-dependent gene misregulation that parallels the onset of RTT-like symptoms in these animals8. b, Mean fold-change in gene expression versus gene length in hippocampus of mice expressing truncated forms of MeCP2 mimicking human disease-causing alleles at four weeks of age. Re-expression of a longer truncated form of MeCP2(G273X) in the MeCP2 KO normalizes expression of long genes more effectively than expression of a shorter truncation of MeCP2(R270X), and parallels the higher degree of phenotypic rescue observed in MeCP2(G273X)-expressing mice compared to MeCP2(R270X)-expressing mice8. c, Mean fold-change in gene expression versus gene length in hippocampus of mice expressing truncated MeCP2 at nine weeks of age. Consistent with the eventual onset of symptoms of these mouse strains, length-dependent gene misregulation is evident in both strains. d, Changes in gene expression for genes binned by length in human MECP2 null ES cells differentiated into neural progenitor cells, neurons cultured for 2 weeks, or neurons cultured for 4 weeks15. In all plots, lines represent mean fold-change in expression for each bin (200 gene bins, 40 gene step), and the ribbon is s.e.m. of genes within each bin. See Methods and Supplementary Table 1 for all sample sizes and other details.
Extended Data Figure 3 High affinity of MeCP2 for mCG, mCA and hmCA in electrophoretic mobility shift assays.
a, Binding of the recombinant methyl-binding domain (MBD) of MeCP2 (amino acids 81–170) to 32P-end-labelled oligonucleotides containing a methylated cytosine in a CA (left) or a CG (right) context competed with unlabelled competitor substituted with unmethylated, methylated, or hydroxymethylated cytosine in a CG or CA context (indicated in bold). Representative full gels showing shifted and unshifted probe in the presence of 50-fold excess of unlabelled competitor (top); close-up of shifted bands over a range of unlabelled competitor (bottom). A mCA-containing oligonucleotide competes for MeCP2 binding with equal or higher efficacy to that of a symmetrically methylated CG oligonucleotide. While hmCG-containing probes compete with similar efficacy to an unmethylated probe, a hmCA-containing probe competes with high efficacy. This difference in affinity of MeCP2 for hmCA- and hmCG-containing probes may explain conflicting results reported for the affinity of MeCP2 for hydroxymethylated DNA18,50,51,52,53(Supplementary Discussion). b, Binding and competition of recombinant MeCP2 MBD (amino acids 78–162, left) or full-length MeCP2 (amino acids 1–486, right) incubated with 32P-end-labelled oligonucleotides containing a methylated cytosine in a CA context and competed with oligonucleotides containing unmethylated, methylated, or hydroxymethylated cytosine in a CG, CA, CT, or CC context. Representative full gels showing 100-fold excess of unlabelled competitor (top); close-up of shifted bands over a range of unlabelled competitor (bottom). The results obtained from competitors containing mCG, mCA, hmCG and hmCA are similar to those shown in a. In addition, both (h)mCT- and (h)mCC-containing oligonucleotides compete for MeCP2 binding with similar efficacy to that of an unmethylated probe. All results shown were observed in at least two independent experiments.
Extended Data Figure 4 ChIP-seq analysis of MeCP2 binding in vivo.
a, Boxplots of input-normalized read density within gene bodies (TSS +3 kb to TTS) for MeCP2 ChIP from the mouse frontal cortex plotted for genes according to quartile of mCA/CA, mCG/CG, hmCA/CA and hmCG/CG in the frontal cortex24 for all genes and genes >100 kb. b, Similar analysis of MeCP2 ChIP from the mouse cortex (left) or cerebellum (right) plotted for genes according to quartile of mCA/CA or mCG/CG for all genes and genes > 100 kb. MeCP2 ChIP-signal is correlated with mCA/CA levels from the frontal cortex, cortex, and cerebellum for all genes and this correlation is more prominent among genes >100 kb. mCG does not show as prominent a correlation with MeCP2 ChIP signal, and hmCG trends towards anti-correlation with MeCP2 ChIP. These results suggest that MeCP2 has a lower affinity for hmCG than mCG, suggesting that, in vivo, hmCG is associated with reduced MeCP2 occupancy (Supplementary Discussion). c, High resolution analysis of high-coverage bisulfite sequencing data from the frontal cortex showing a correlation between MeCP2 ChIP signal and mCA. Input-normalized ChIP signal plotted for mCA levels for 500-bp bins tiled across all genes. d, Aggregate plots of MeCP2 input-normalized ChIP signal (top) and relative methylation (log2 enrichment in mC as compared to the flanking regions) for mCA, mCC, mCT, and mCG (bottom) are plotted around the 31,479 summits of MeCP2 ChIP enrichment identified using the MACS peak-calling algorithm40 (red) or 31,479 randomly selected control sites (grey, see Methods). See Methods and Supplementary Table 1 for sample sizes and other details.
Extended Data Figure 5 Genomic analysis of mCG, hmCG, and hmCA in length-dependent gene regulation by MeCP2.
a–c, Mean methylation of CG dinucleotides (mCG/CG) within gene bodies (transcription start site +3 kb, up to transcription termination site) in the cortex (a), hippocampus (b) and cerebellum (c) for genes binned according to length. d–f, Mean fold-change in gene expression in MeCP2 KO compared to wild type in the cortex (d), hippocampus (e), and cerebellum (f) for genes binned according to mCG levels (mCG/CG) within gene bodies. g, Mean hmCG levels (hmCG/CG) within gene bodies in the frontal cortex24 for genes binned according to length. h, Mean fold-change in gene expression in MeCP2 KO compared to wild type for genes binned according to hmCG levels (hmCG/CG) within gene bodies in the frontal cortex24 i, Mean hmCA levels (hmCA/CA) within gene bodies in the frontal cortex24 for genes binned according to length. j, Mean fold-change in gene expression in MeCP2 KO compared to wild type genes binned according to hmCA levels (hmCA/CA) within gene bodies in the frontal cortex24. In all panels, mean values for each bin are indicated as a line (200 gene bins, 40 gene step); ribbon depicts s.e.m. for genes within each bin. See Methods and Supplementary Table 1 for sample sizes and other details.
Extended Data Figure 6 Genomic analysis supports a role for mCA in length-dependent gene regulation by MeCP2.
a–c, Mean methylation at CA dinucleotides (mCA/CA) within gene bodies (TSS +3 kb to TTS) in cortex (a), hippocampus (b), and cerebellum (c) for genes binned by length. d–f, Mean changes in gene expression in cortex (d), hippocampus (e), and cerebellum (f) of MeCP2 KO for high mCA genes (top 25% mean gene body mCA/CA) and low mCA genes (bottom 66% mean gene body mCA/CA) binned by length. g–i, Mean changes in gene expression in cortex (g), hippocampus (h), and cerebellum (i) of MeCP2 KO for genes binned according to average gene body mCA/CA levels. j–l, Mean changes in gene expression in cortex (j), hippocampus (k), and cerebellum (l) of MeCP2 KO mice for long genes (top 25%) and short genes (bottom 25%) in each brain region binned by gene body mCA/CA level. A correlation between fold-change in the MeCP2 KO and mCA/CA for all genes is less prominent, or not observed, in the hippocampus and cerebellum for all genes together (h, i), but it is clear for the longest genes in the genome analysed alone (k, l). Note that average levels of mCA appear lower in hippocampus and cerebellum compared to cortex (compare y axis in a, b and c), and may explain why a correlation across all genes in not detected in these brain regions. In long genes analysed alone the cumulative effect of higher mCA levels and integration across the gene may be larger, resulting in a detectable effect. In all panels, the line indicates the mean for 200 gene bins, with a 40 gene step; ribbon depicts s.e.m. for genes within each bin. Note that, for completeness, data from analysis of the cortex presented in Fig. 2 are re-presented here. See Methods and Supplementary Table 1 for sample sizes and other details.
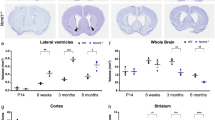
Extended Data Figure 7 Conditional knockout of Dnmt3a in vivo.
a, Diagram of the Dnmt3a locus and Cre-dependent conditional knockout strategy for Dnmt3a26. LoxP sites (green triangles) flank exon 17, which is removed following Cre-mediated recombination. Primers (purple arrows) were designed to flank exons 17 and 18. The wild-type (WT), floxed (FLX), and knockout (KO) allele are depicted. b, Representative PCR genotyping for tail DNA samples indicates presence or absence of the floxed (flx, ∼800 bp), wild-type (WT, ∼750 bp), and knockout (KO, ∼500 bp) alleles. Separate genotyping reaction for the Nestin-cre transgene (∼250 bp) is shown. c, Efficient excision of the floxed exon is detected in cerebellar DNA from conditional knockout (Dnmt3aflx/flx; Nestin-Cre+/−, Dnmt3a cKO) mice but not from and control animals (Dnmt3aflx/flx, Control). d, Western blot analysis of Dnmt3a, MeCP2, and Gapdh (loading control) protein from the cerebellum of control and Dnmt3a cKO adult mice. All results shown were observed in at least two independent experiments.
Extended Data Figure 8 Analysis of MeCP2-repressed genes and FMRP target genes.
a, Mean fold-change in mRNA expression for examples of MeCP2-repressed genes across three different Mecp2 mutant genotypes (KO, OE, and R306C) and six brain regions. P values for each gene are derived from the mean z scores for fold-change across all data sets (see Methods). b, Gene expression and CA methylation data from the cerebellum for selected MeCP2-repressed genes from a (right), as well as examples of extremely long genes (>100 kb) that are not enriched for mCA and are not misregulated (left). Fold-changes in mRNA expression in Mecp2 mutants and the Dnmt3a cKO are shown (left axis), as well as mean mCA levels (grey; right axis). Red line indicates genomic median for gene body mCA/CA c, Boxplots of mCA levels in MeCP2-repressed genes compared to all genes. d, Mean fold-change for MeCP2-repressed genes in eight ‘training data sets’ used to define these genes (see Methods), and nine ‘test data sets’: three Mecp2 mutant data sets not used to define MeCP2-repressed genes (CTX MeCP2 KO and CB MeCP2(R306C), generated in this study; HC MeCP2 KO 4 week, analysed from Baker et al.8), and six data sets from brains of mouse models of neurological dysfunction generated using the same microarray platforms as the MeCP2 data sets (GEO accession numbers in order: GSE22115, GSE27088, GSE43051, GSE47706, GSE44855, GSE52584). Error bars are s.e.m. of MeCP2–repressed gene expression across samples (n = 4–8 microarrays per genotype per data set); P < 0.01, one-tailed t-test, Benjamini–Hochberg correction. Note that significance testing was not performed on training data sets. Brain regions indicated as in Fig. 1, (WB, whole brain). e, Cumulative distribution function (CDF) of gene lengths plotted exclusively for genes that are among the top 60% of expression levels in the brain (Supplementary Discussion). The extreme length of MeCP2-repressed genes and genes encoding FMRP target mRNAs29 when controlling for expression level indicates that the long length of these genesets is not a secondary effect of the preferential expression of long genes in the brain (P < 1 × 10−15 for each geneset versus all expressed genes; two-sample Kolmogorov–Smirnov test). f, The CDF of gene lengths for all genes compared to an independent set of FMRP targets identified by Brown and colleagues45 (P < 1 × 10−15, Kolmogorov–Smirnov-test). g, CDF of gene lengths for genes expressed at similar levels in the brain and other somatic tissues (Supplementary Discussion). The extreme length of each geneset (P < 1 × 10−15, Kolmogorov–Smirnov test) when filtering for genes that are expressed in all tissues indicates that regulation of long genes by MeCP2 and FMRP is not dependent on brain-specific expression. h, CDF of mature mRNA lengths for MeCP2-repressed genes, and FMRP target genes (P < 1 × 10−11 for each geneset versus all genes, Kolmogorov–Smirnov test). i, Overlap of MeCP2-repressed genes and putative FMRP target mRNAs29 (P < 5 × 10−5, hypergeometric test). Expected overlap was calculated by dividing the expected overlap genome-wide (hypergeometric distribution) according to the distribution of all gene lengths in the genome. See Methods and Supplementary Table 1 for sample sizes and other details.
Extended Data Figure 9 Consequences of long gene misregulation in neurons.
a, Mean expression of genes binned according to length in human neural and non-neural tissues. Mean expression for genes within each bin (200 gene bins, 40 gene step) is indicated by the line; ribbon represents the s.e.m. of genes within each bin. b, Western blot analysis of MeCP2 from primary cortical neurons after control or MeCP2 shRNA knockdown (KD) and treatment with DMSO vehicle (−) or topotecan (+). c, Heatmap summary of nCounter analysis for the expression of selected MeCP2-repressed (MR) genes from primary neurons treated with control or MeCP2 shRNA and topotecan (n = 3–4). Normalized log2 fold-change relative to the DMSO-treated, control KD is shown. MeCP2 KD conditions are significantly different from control, (P = 1 × 10−4, repeated measures ANOVA across 8 genes). Newman–Keuls corrected, post-hoc comparisons: P < 0.05 control KD, 0 nM drug versus MeCP2 KD, 0 nM drug; P > 0.05, control KD, 0 nM drug versus MeCP2 KD, 50 nM drug; P < 0.05 MeCP2 KD, 0 nM drug versus MeCP2 KD, 50 nM drug. d, Bioanalyzer profiles of 18S and 28S ribosomal RNA (top) and total RNA quantification (bottom) for treated neurons (n = 3–5). Total RNA values normalized to DMSO-treated control KD, red dashed line. Two-way repeated measures ANOVA indicates a significant effect of KD (P < 0.01) and drug treatment (P < 0.05). Rescue assessed by one-tailed t-test, Bonferroni multiple testing correction, P < 0.05.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and additional references. (PDF 159 kb)
Supplementary Table 1
This file contains the read numbers and statistics for bisulfite-seq analysis of wild type cerebellum and cortex. (XLSX 20 kb)
Supplementary Table 2
This fie contains the primers used for quantitative RT-PCR analysis. (XLSX 43 kb)
Supplementary Table 3
This file contains Identification and analysis of MeCP2-repressed genes. (XLSX 2237 kb)
Rights and permissions
About this article
Cite this article
Gabel, H., Kinde, B., Stroud, H. et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 (2015). https://doi.org/10.1038/nature14319
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14319
This article is cited by
-
Overcoming genetic and cellular complexity to study the pathophysiology of X-linked intellectual disabilities
Journal of Neurodevelopmental Disorders (2024)
-
Circular RNAs arising from synaptic host genes during human neuronal differentiation are modulated by SFPQ RNA-binding protein
BMC Biology (2023)
-
Buffering of transcription rate by mRNA half-life is a conserved feature of Rett syndrome models
Nature Communications (2023)
-
Epigenetic and epitranscriptomic regulation of axon regeneration
Molecular Psychiatry (2023)
-
Methylation across the central dogma in health and diseases: new therapeutic strategies
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.