Abstract
Wing polyphenism is an evolutionarily successful feature found in a wide range of insects1. Long-winged morphs can fly, which allows them to escape adverse habitats and track changing resources, whereas short-winged morphs are flightless, but usually possess higher fecundity than the winged morphs1,2,3. Studies on aphids, crickets and planthoppers have revealed that alternative wing morphs develop in response to various environmental cues1,2,4,5,6,7,8, and that the response to these cues may be mediated by developmental hormones, although research in this area has yielded equivocal and conflicting results about exactly which hormones are involved4,8,9,10. As it stands, the molecular mechanism underlying wing morph determination in insects has remained elusive. Here we show that two insulin receptors in the migratory brown planthopper Nilaparvata lugens, InR1 and InR2, have opposing roles in controlling long wing versus short wing development by regulating the activity of the forkhead transcription factor Foxo. InR1, acting via the phosphatidylinositol-3-OH kinase (PI(3)K)–protein kinase B (Akt) signalling cascade, leads to the long-winged morph if active and the short-winged morph if inactive. InR2, by contrast, functions as a negative regulator of the InR1–PI(3)K–Akt pathway: suppression of InR2 results in development of the long-winged morph. The brain-secreted ligand Ilp3 triggers development of long-winged morphs. Our findings provide the first evidence of a molecular basis for the regulation of wing polyphenism in insects, and they are also the first demonstration—to our knowledge—of binary control over alternative developmental outcomes, and thus deepen our understanding of the development and evolution of phenotypic plasticity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
The cDNA sequences of NlInR1 and NlInR2 have been deposited in GenBank under accession numbers KF974333 and KF974334, respectively. Gene sequences used for dsRNA synthesis have been deposited in GenBank under the following accession numbers: KF974335 (NlChico), KF974336 (NlLnk), KF974337 (NlAkt), KF974338 (NlPten), KF974339 (NlFoxo), KF974340–KF974343 (NlIlp1–4), KF974348 (NlErk), KF974349 (NlRaf), KF974350 (NlTor), KM099280 (NlRaptor), KM099281 (NlRheb), KF974344 (SfInR1), KF974345 (SfInR2), KF974346 (LsInR1) and KF974347 (LsInR2).
References
Zera, A. J. in Phenotypic Plasticity of Insects: Mechanisms and Consequences (eds Whitman, D. W. & Ananthakrishnan, T. N. ) 609–653 (Science, 2009)
Zera, A. J. & Denno, R. F. Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol. 42, 207–230 (1997)
Zhao, Z. & Zera, A. J. Differential lipid biosynthesis underlies a tradeoff between reproduction and flight capability in a wing-polymorphic cricket. Proc. Natl Acad. Sci. USA 99, 16829–16834 (2002)
Denno, R. F., Douglas, L. W. & Jacobs, D. Crowding and host plant nutrition: environmental determinants of wing-form in Prokelisia marginata. Ecology 66, 1588–1596 (1985)
Nijhout, H. F. Control mechanisms of polyphenic development in insects. Bioscience 49, 181–192 (1999)
Braendle, C., Friebe, I., Caillaud, M. C. & Stern, D. L. Genetic variation for an aphid wing polyphenism is genetically linked to a naturally occurring wing polymorphism. Proc. R. Soc. Lond. B 272, 657–664 (2005)
Roff, D. A. & Fairbairn, D. J. The evolution and genetics of migration in insects. Bioscience 57, 155–164 (2007)
Simpson, S. J., Sword, G. A. & Lo, N. Polyphenism in insects. Curr. Biol. 21, R738–R749 (2011)
Lees, A. D. Action of juvenile hormone mimics on the regulation of larval–adult and alary polymorphism in aphids. Nature 267, 46–48 (1977)
Braendle, C., Davis, G. K., Brisson, J. A. & Stern, D. L. Wing dimorphism in aphids. Heredity 97, 192–199 (2006)
Kisimoto, R. Effect of crowding during the larval period on the determination of the wing-form of an adult plant-hopper. Nature 178, 641–642 (1956)
Iwanaga, K., Tojo, S. & Nagata, T. Immigration of the brown planthopper, Nilaparvata lugens, exhibiting various responses to density in relation to wing morphism. Entomol. Exp. Appl. 38, 101–108 (1985)
Iwanaga, K. & Tojo, S. Effects of juvenile hormone and rearing density on wing dimorphism and oöcyte development in the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 32, 585–590 (1986)
Syobu, S., Mikuriya, H., Yamaguchi, J., Matsuzaki, M. & Matsumura, M. Fluctuations and factors affecting the wing-form ratio of the brown panthopper, Nilaparvata lugens Stal in rice fields. Jap. J. Appl. Entomol. Zool. 46, 135–143 (2002)
Hartfelder, K. & Emlen, D. in Insect Endocrinology (ed. Gilbert, L. I. ) 464–522 (Elsevier, 2012)
Brogiolo, W. et al. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221 (2001)
Clancy, D. J. et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104 (2001)
Nakae, J., Kido, Y. & Accili, D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 22, 818–835 (2001)
Oldham, S. & Hafen, E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13, 79–85 (2003)
Edgar, B. A. How flies get their size: genetics meets physiology. Nature Rev. Genet. 7, 907–916 (2006)
Taniguchi, C. M., Emanuelli, B. & Kahn, C. R. Critical nodes in signalling pathways: insights into insulin action. Nature Rev. Mol. Cell Biol. 7, 85–96 (2006)
Hietakangas, V. & Cohen, S. M. Regulation of tissue growth through nutrient sensing. Annu. Rev. Genet. 43, 389–410 (2009)
Teleman, A. A. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425, 13–26 (2010)
Marshall, L., Rideout, E. J. & Grewal, S. S. Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J. 31, 1916–1930 (2012)
Britton, J. S., Lockwood, W. K., Li, L., Cohen, S. M. & Edgar, B. A. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239–249 (2002)
Cantley, L. C. The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 (2002)
Ogg, S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 (1997)
Puig, O., Marr, M. T., Ruhf, M. L. & Tjian, R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 (2003)
Maehama, T. & Dixon, J. E. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 9, 125–128 (1999)
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006)
Xue, J. et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15, 521 (2014)
Xu, H. J. et al. Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 22, 635–647 (2013)
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001)
Parrou, J. L. & Francois, J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248, 186–188 (1997)
Xue, J. et al. Molecular characterization of the flightin gene in the wing-dimorphic planthopper, Nilaparvata lugens, and its evolution in Pancrustacea. Insect Biochem. Mol. Biol. 43, 433–443 (2013)
Hirumi, H. & Maramorosch, K. in Invertebrate Tissue Culture (ed. Vago, C. ) 307–337 (Academic, 1971)
Géminard, C., Rulifson, E. J. & Léopold, P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10, 199–207 (2009)
Acknowledgements
We thank R.-Z. Zhang for help with BPH imaging (Fig. 1a). This work was supported by the National Basic Research Program of China (973 Program, no. 2010CB126205) and by the National Science Foundation of China (no. 31201509 and no. 31471765).
Author information
Authors and Affiliations
Contributions
H.-J.X. conceived and designed the study, wrote the paper, helped perform experiments and analysed the data. J.X. performed most experiments and helped with data analysis. B.L., Y.-Q.J., Q.L., S.-F.H. and J.-Y.X. helped perform experiments and antibody preparation. X.-C.Z. and J.-C.Z. performed gene cloning and immunoprecipitation. X.-F.M. performed RACE experiments. H.-W.F., Y.-X.Y. and P.-L.P. performed qRT–PCR. Y.-Y.B. and H.F.N. discussed data and revised the manuscript. C.-X.Z. organized and directed the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 cDNA sequences of NlInR1 and NlInR2.
a, b, NlInR1 (a) and NlInR2 (b) share highly similar domain architectures. The signal peptide (SP) indicated by a vertical arrow is produced via a cleavage site at the amino terminus. Two ligand-binding loops (L1 and L2) and the furin-like cysteine-rich (Fu) region are underlined with solid lines. Three fibronectin type 3 (FN3) domains are labelled with dashed underlines. A single transmembrane (TM) region is highlighted with a box. An ‘NPXY’ motif is shown in green. The highly conserved tyrosine kinase domain (TyrKc) is indicated in blue. A triple tyrosine cluster (YXXXYY), is indicated in red. c, The sequence identity (%) of each domain in the InR receptors compared to its counterpart in the human insulin receptor (HsInR). Dm, D. melanogaster. d, The alignment of the N-terminal part of the Fu domains of NlInR1, NlInR2, DmInR and HsInR. The four cysteine residues that are absent in NlInR2 are indicated by asterisks.
Extended Data Figure 2 Spatio-temporal expression analyses of NlInR1 and NlInR2.
a, Quantitative polymerase chain reaction with reverse transcription (qRT–PCR) analysis was performed on various tissues dissected from 4th-instar nymphs (n = 100), 5th-instar nymphs (n = 60) and adult females (n = 30). a–c, NlInR1 was widely expressed in all tissue samples collected from 4th-instar nymphs (a), 5th-instar nymphs (b) or adult females (c). d–f, NlInR2 was predominately expressed in wing buds in 4th-instar nymphs (d) and 5th-instar nymphs (e), but was widely expressed in adult tissues (f). a–f, Error bars represent the s.e.m of three technical replicates. g, Relative expression levels of NlInR1 and NlInR2 across development. First-instar nymphs (n = 60), 2nd-instar nymphs (n = 60), 3rd-instar nymphs (n = 20), 4th-instar nymphs (n = 20), 5th-instar (n = 20) nymphs and adults (n = 20) were used for RNA extraction. A relatively high expression of NlInR2 was observed in the 4th- and 5th-instar nymphs. Mean ± s.e.m. from three experiments, P values are indicated (Student’s t-test). SWF, short-winged females; LWF, long-winged females; SWM, short-winged males; LWM, long-winged males.
Extended Data Figure 3 Effects of knockdown of components of the IIS pathway.
a, Knockdown of NlInR2, NlFoxo or NlPten. b, Knockdown of NlInR1, NlChico or NlAkt. Forewing veins are patterned normally. c, The proportion of short-winged (SW) BPHs, from three experiments (mean ± s.e.m.). dsgfp (n = 142 females, 161 males), dsNlChico (n = 86 females, 98 males), dsNlLnk (n = 100 females, 117 males), dsNlPten (n = 89 females, 78 males), dsNlAkt (n = 106 females, 89 males) and dsNlFoxo (n = 78 females, 82 males). P < 0.05 and P ≤ 0.001 (Student’s t-test), difference from dsgfp. Source data are provided in Supplementary Data 4.
Extended Data Figure 4 Validation of RNAi effect using the second dsRNA molecules, and knockdown of NlInRs in BPHs at 3rd- and 4th-instar nymphs.
a, The proportion of short-winged (SW) BPHs after treatment of 2nd-instar nymphs with the second dsRNAs targeting NlInRs (dsNlInR1_B or dsNlInR2_B). dsgfp (n = 124 females, 100 males), dsNlInR1_B (n = 90 females, 92 males) and dsNlInR2_B (n = 103 females, 116 males). b, Knockdown of NlInRs in 3rd-instar nymphs. dsgfp (n = 117 females, 114 males), dsNlInR1 (n = 109 females, 130 males) and dsNlInR2 (n = 90 females, 100 males). c, Knockdown of NlInRs in 4th-instar nymphs. dsgfp (n = 101 females, 126, males), dsNlInR1 (n = 90 females, 117 males) and dsNlInR2 (n = 94 females, 100 males). d, The 3rd-instar nymphs were treated with a second dsRNA targeting several key components in the canonical insulin signalling pathway. dsgfp (n = 96 females, 100 males), dsNlChico_B (n = 72 females, 78 males), dsNlAkt_B (n = 86 females, 78 males), dsNlPten_B (n = 111 females, 88 males) and dsNlFoxo_B (n = 95 females, 91 males). a–d, Mean ± s.e.m. from three experiments. P < 0.05 and P ≤ 0.001 (Student’s t-test), difference from the ratio obtained for dsgfp. Source data are provided in Supplementary Data 5.
Extended Data Figure 5 Knockdown of components of the TORC1 and the Ras/MAPK signalling cascades have no effect on wing morph switch.
a, Third-instar nymphs were treated with dsRNAs targeting NlTor and NlRaptor genes in the TORC1 complex, and targeting NlErk and NlRaf in the Ras/MAPK signalling pathway. dsgfp (n = 134 females, 153 males), dsNlTor (n = 122 females, 130 males), dsNlRaptor (n = 103 females, 95 males), dsNlErk (n = 138 females, 148 males) and dsNlRaf (n = 148 females, 143 males). Mean ± s.e.m. from three experiments, no significant change from dsgfp (Student’s t-test). b, Knockdown of NlRheb, an activator for TORC1 signalling activity. Third-instar nymphs were used for dsNlRheb treatment. Seventy-eight individuals (74%) failed to extricate themselves from the old cuticle, and died without completing ecdysis. Twenty-eight individuals finished nymph–adult transformation, of which twenty-five individuals (24%) were unable to stretch their wings correctly, and only three individuals (3%) showed normal morphology. Source data are provided in Supplementary Data 6.
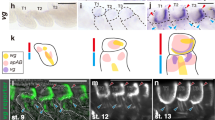
Extended Data Figure 6 NlInR2 regulates wing morph switch in a tissue-specific way.
a–n, Second-instar nymphs were treated with dsRNAs targeting gfp, NlInR1, NlInR2, NlChico or NlFoxo, and 4th-instar nymphs were used for the dsNlAkt treatment. a, The short-winged BPHs (dsgfp-SW, n = 20) and long-winged BPHs (dsgfp-LW, n = 20) had a similar length of hind tibiae. The BPHs treated either with dsNlInR2 (n = 20) or with double RNAi (dsNlInR2;dsNlFoxo, n = 20) had a similar length of hind tibiae as the dsgfp-LW (n = 20). b, The BPHs treated with dsNlInR2 (n = 20) or dsNlInR2;dsNlFoxo (n = 20) possessed forewings of the same size or only slightly smaller than those of dsgfp-BPHs (dsgfp-LW, n = 20). c, d, Knockdown of NlInR1 (n = 20), NlChico (n = 20) or NlAkt (n = 20) further reduced the hind tibiae length (c) and forewing size (d) compared to short-winged BPHs treated with dsgfp (dsgfp-SW, n = 20). e, Knockdown of NlInR1 (n = 50) or NlChico (n = 44) but not NlInR2 (n = 66) delayed nymphal development. f, Knockdown of NlInR1 but not NlInR2 resulted in body weight loss in 5th-instar nymphs. dsgfp (n = 107; 110, 24 h; 72 h), dsNlInR1 (n = 117; 82, 24 h; 72 h), and dsNlInR2 (n = 111; 103, 24 h; 72 h). ‘24 h’ and ‘72 h’ represent 24 h and 72 h after ecdysis, respectively. g–l, Knockdown of NlInR1 but not NlInR2 reduced levels of glycogen, trehalose and glucose both in nymphs (g–i) and adult females (j–l). m, n, Knockdown of NlInR1 but not NlInR2 increased levels of triglycerides in both nymphs (m) and adult females (n). a–n, Mean ± s.e.m. from three experiments. Tukey’s test in a, b, and Student’s t-test in c–n, difference from dsgfp (P < 0.05 and P ≤ 0.001). Source data are provided in Supplementary Data 7.
Extended Data Figure 7 Subcellular localization of NlFoxo in wing buds and fat body.
a, b, Localization of NlFoxo in wing buds (a) and fat body (b). Third-instar nymphs were treated with dsRNAs, and wing buds and fat body were dissected from 5th-instar nymphs (48 h after ecdysis). For LY294002 treatment, wing buds and fat body dissected from untreated 5th-instar nymphs (48 h after ecdysis) were used for immuno-staining. Red, anti-NlFoxo. Blue (DAPI), cell nucleus. c, Proportion of cells with DAPI/Foxo co-localization in wing buds and fat body treated with dsNlInR2 or dsNlPten. Error bars represent mean ± s.e.m. from cells in three images. P ≤ 0.001 (Student’s t-test). d, Knockdown of NlInR2 did not increase P-NlAkt levels in the fat body. Fat body treated with dsgfp, dsNlInR1, dsNlInR2 or dsNlPten was probed with various antibodies.
Extended Data Figure 8 Tissue distribution of NlIlp1–4 in nymphs and adult females.
a–l, Various tissues dissected from 4th-instar nymphs (n = 100), 5th-instar nymphs (n = 60) and adult females (n = 30) were exposed to qRT–PCR assays to detect NlIlp1 (a–c), NlIlp2 (d–f), NlIlp3 (g–i) and NlIlp4 (j–l) transcripts. The fold changes for transcripts in the head versus the fat body are indicated. Mean ± s.e.m. from three technical replicates.
Extended Data Figure 9 Knockdown of two insulin receptors in planthoppers, Sogatella furcifera and Laodelphax striatellus.
a, The wing morphs of S. furcifera planthoppers (SFPs). b, Knockdown of S. furcifera (Sf)InR1 resulted in the short-winged morph. dsgfp (n = 110 females, 89 males), dsSfInR1 (n = 101 females, 135 males) and dsSfInR2 (n = 132 females, 143 males). c, The wing morphs of L. striatellus planthoppers (LSPs). d, Knockdown of L. striatellus (Ls)InR2 resulted in the long-winged morph. dsgfp (n = 107 females, 123 males), dsLsInR1 (n = 42 females, 128 males) and dsLsInR2 (n = 113 females, 107 males). e, Third-instar nymphs were treated with dsRNAs targeting gfp (n = 200), LsInR1 (n = 180) or LsInR2 (n = 190). The survival rate of LSPs was monitored every day before metamorphosis. Mean ± s.e.m. from three experiments, no significant change from dsgfp (Student’s t-test). f, The metamorphosis rate was monitored every day at nine days after dsRNAs treatments. dsgfp (n = 200), dsLsInR1 (n = 180) and dsLsInR2 (n = 190). Most dsLsInR1-treated nymphs (>70%) died before adult eclosion, and thus yielded a significantly low metamorphosis rate. b–f, Mean ± s.e.m. from three experiments. P < 0.05 and P ≤ 0.001 (Student’s t-test), difference from dsgfp. Source data are provided in Supplementary Data 8.
Extended Data Figure 10 Examination of RNAi efficiency by qRT–PCR.
a–v, Individual nymphs were pooled (n = 20) to extract total RNA after dsRNAs treatments, and cDNA was synthesized with random primers. The relative expression of each gene was normalized to the expression level of ribosomal 18S rRNA. Mean ± s.e.m. from three experiments.
Supplementary information
Supplementary information
This file contains Supplementary notes 1-9 and Supplementary References. (PDF 800 kb)
Supplementary Data
This file contains Supplementary Table 1 which shows the sex ratio of BPHs following dsNlInRs treatments. (XLSX 9 kb)
Supplementary Data
This file contains Supplementary Table 2, a list of the main primers used in this study. (XLS 40 kb)
An immunofluorescence assay of NlILP3 in brains of 5th-instar nymphs
The stained nerve cords (red) were crossed over a short distance from the medial neurosecretory cells to extend backwards in an arc through the brain. The cell nucleus is stained with DAPI (blue). (MOV 1542 kb)
Rights and permissions
About this article
Cite this article
Xu, HJ., Xue, J., Lu, B. et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467 (2015). https://doi.org/10.1038/nature14286
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14286
This article is cited by
-
Histone deacetylases regulate organ-specific growth in a horned beetle
EvoDevo (2024)
-
Salivary proteins potentially derived from horizontal gene transfer are critical for salivary sheath formation and other feeding processes
Communications Biology (2024)
-
Genetic diversity of RNA viruses infecting invertebrate pests of rice
Science China Life Sciences (2024)
-
Long-wave opsin involved in body color plastic development in Nilaparvata lugens
BMC Genomics (2023)
-
Planthopper salivary sheath protein LsSP1 contributes to manipulation of rice plant defenses
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.