Abstract
In Drosophila, rapid temperature changes are detected at the periphery by dedicated receptors forming a simple sensory map for hot and cold in the brain1. However, flies show a host of complex innate and learned responses to temperature, indicating that they are able to extract a range of information from this simple input. Here we define the anatomical and physiological repertoire for temperature representation in the Drosophila brain. First, we use a photolabelling strategy2 to trace the connections that relay peripheral thermosensory information to higher brain centres, and show that they largely converge onto three target regions: the mushroom body, the lateral horn (both of which are well known centres for sensory processing) and the posterior lateral protocerebrum, a region we now define as a major site of thermosensory representation. Next, using in vivo calcium imaging3, we describe the thermosensory projection neurons selectively activated by hot or cold stimuli. Fast-adapting neurons display transient ON and OFF responses and track rapid temperature shifts remarkably well, while slow-adapting cell responses better reflect the magnitude of simple thermal changes. Unexpectedly, we also find a population of broadly tuned cells that respond to both heating and cooling, and show that they are required for normal behavioural avoidance of both hot and cold in a simple two-choice temperature preference assay. Taken together, our results uncover a coordinated ensemble of neural responses to temperature in the Drosophila brain, demonstrate that a broadly tuned thermal line contributes to rapid avoidance behaviour, and illustrate how stimulus quality, temporal structure, and intensity can be extracted from a simple glomerular map at a single synaptic station.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gallio, M., Ofstad, T. A., Macpherson, L. J., Wang, J. W. & Zuker, C. S. The coding of temperature in the Drosophila brain. Cell 144, 614–624 (2011)
Ruta, V. et al. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature 468, 686–690 (2010)
Wang, J. W., Wong, A. M., Flores, J., Vosshall, L. B. & Axel, R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282 (2003)
Ito, K. et al. A systematic nomenclature for the insect brain. Neuron 81, 755–765 (2014)
Datta, S. R. et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 452, 473–477 (2008)
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnol. 22, 1567–1572 (2004)
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012)
Feinberg, E. H. et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008)
Gordon, M. D. & Scott, K. Motor control in a Drosophila taste circuit. Neuron 61, 373–384 (2009)
Nicolai, L. J. et al. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc. Natl Acad. Sci. USA 107, 20553–20558 (2010)
Zhang, Y. Q., Rodesch, C. K. & Broadie, K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis 34, 142–145 (2002)
Baines, R. A., Uhler, J. P., Thompson, A., Sweeney, S. T. & Bate, M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21, 1523–1531 (2001)
Thorne, N. & Amrein, H. Atypical expression of Drosophila gustatory receptor genes in sensory and central neurons. J. Comp. Neurol. 506, 548–568 (2008)
Ni, L. et al. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature 500, 580–584 (2013)
Benton, R., Vannice, K. S., Gomez-Diaz, C. & Vosshall, L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009)
Karuppudurai, T. et al. A hard-wired glutamatergic circuit pools and relays UV signals to mediate spectral preference in Drosophila. Neuron 81, 603–615 (2014)
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013)
Stocker, R. F., Lienhard, M. C., Borst, A. & Fischbach, K. F. Neuronal architecture of the antennal lobe in Drosophila melanogaster. Cell Tissue Res. 262, 9–34 (1990)
Tanaka, N. K., Endo, K. & Ito, K. Organization of antennal lobe-associated neurons in adult Drosophila melanogaster brain. J. Comp. Neurol. 520, 4067–4130 (2012)
Bruckner, S. et al. BrainGazer–visual queries for neurobiology research. IEEE Trans. Vis. Comput. Graph. 15, 1497–1504 (2009)
Lai, J. S., Lo, S. J., Dickson, B. J. & Chiang, A. S. Auditory circuit in the Drosophila brain. Proc. Natl Acad. Sci. USA 109, 2607–2612 (2012)
Wang, Z., Singhvi, A., Kong, P. & Scott, K. Taste representations in the Drosophila brain. Cell 117, 981–991 (2004)
Acknowledgements
We thank V. Ruta and B. Noro for providing PA-GFP flies and reagents, and especially G. Rubin for providing access to the FlyLight collection before publication. We are grateful to N. Ryba, D. Yarmolinsky, C. Zuker and members of the Gallio laboratory for critical comments on the manuscript. Z. Turan and A. Kuang provided technical assistance and we thank C. Ulmer and a number of undergraduate students for fly husbandry. This work was supported by NIH grant 1R01NS086859-01 (to M.G.) and by training grant 2T32MH067564 (to D.D.F.).
Author information
Authors and Affiliations
Contributions
M.G. and D.D.F. designed the study, carried out the imaging experiments, analysed data (with help from G.C.J.), and wrote the paper; D.D.F., G.C.J. and M.G. ran and analysed all behavioural experiments; P.J.K. carried out GRASP experiments using transgenic lines produced by M.G. and L.J.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
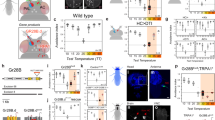
Extended Data Figure 1 Targeted photo-conversion of PA-GFP reveals candidate tPN pathways.
a, Diagram of the fly brain showing the location of the PAL. b, Schematic diagram showing the expected location of ‘cold’ and ‘hot’ glomeruli (blue and red, respectively). c–e, The hot glomerulus is visualized by tdTomato, driven by Gr28b.d-LexA (red). e, Targeted photo-activation of pan-neuronal PA-GFP (syb-Gal4/UAS-SPA) reveals ascending pathways (white arrowheads). Panels d and e are imaged under the same conditions, before (d) and after (e) photoconversion. MB, mushroom body; PAL, posterior antennal lobe; SEZ, sub oesophageal zone; AL, antennal lobe; AN, antennal nerve; SPP, super peduncular protocerebrum. Scale bars, 25 μm.
Extended Data Figure 2 Synaptic-targeted GFP reconstitution across synaptic partners (GRASP) reveals specific connectivity patterns between antennal temperature receptors and tPN classes.
Using an n-synaptobrevin fragment to target spGFP1-10 to the pre-synaptic membrane (see Methods) increases the specificity of GRASP fluorescence at synapses between thermosensory neurons and projection neurons in the PAL. a, Immunofluorescence staining of GRASP contacts between hot and cold receptor neurons (IR94a-LexA, Aop-CD4:spGFP1-10) and projection neurons (VT46265, UAS-CD4:spGFP11) shows diffuse expression of spGFP1-10 throughout the membranes of the hot and cold receptor neurons (red), in addition to the GRASP signal at the synapses between hot cells and VT46265 projection neurons (green). Some non-synaptic GRASP fluorescence is seen at sites of contact between the receptor neurons and fibres of passage (arrow in a, and see cartoon below for illustration). In contrast, synaptically targeted syb–GRASP (or syb:GRASP) (b) shows punctate expression of syb-spGFP1-10 (red) and specific GRASP signal (green) only at the hot-cell synapses of the PAL. c, Representative native GRASP fluorescence between either (d, f, h) hot and cold temperature receptors (IR94a-LexA; Aop-CD4:spGFP1-10) or hot only temperature receptors (e, g, i, GR28b.d-LexA; Aop-syb-spGFP1-10) onto three projection neuron types (VT40053, R60H12 and VT46265, each driving UAS-CD4:spGFP11). GRASP connectivity corresponds well to their tuning (see text and Extended Data Table 1 for details). Scale bars in d–i, 25 μm.
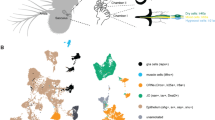
Extended Data Figure 3 Target regions of tPN cell types.
Transgenic expression of synaptotagmin–GFP reveals the location of putative pre-synaptic termini for each of the identified tPN cell types. a, Three-dimensional reconstruction of key target regions (the mushroom body and lateral horn, based on nc82 staining) showing their relationship to the tPN pathways (colour and abbreviations are as in Fig. 1; note that the boundaries of the PLP are not clearly demarcated by nc82 staining, hence the PLP was not reconstructed but outlined; see Methods for details). b, d, f, h, j, l, Three-dimensional reconstructions of specific tPN pathways; c, e, g, i, k, m, two-photon stacks of brains expressing synaptotagmin–GFP under the control of Gal4 drivers expressed in neurons innervating such pathways (the driver name appears at the bottom of each panel). GFP fluorescence identifies putative pre-synaptic structures in the target regions (arrowheads). In c, e and i, the mushroom body calyx is circled. Scale bars, 25 μm.
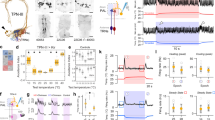
Extended Data Figure 4 Temporal dynamics of slow- and fast-adapting cold-activated tPNs.
Calcium transients of slow- and fast-adapting cold-activated tPNs differ significantly in the speed of decay. a, The response of a fast tPN (right plot, grey trace) returns to baseline much more rapidly than that of a slow one (left plot, grey trace). In a, a small cooling stimulus and the response it elicited in a representative cell are normalized and overlaid to facilitate comparison of dynamics (note that the sign of cooling stimuli is inverted for clarity, ΔT ≈ −3 °C, responses are ΔF/F recorded at cell bodies, stimuli and responses are recorded synchronously; see Methods for details). The decay constant of each response is shown next to each plot (τ, defined as the time from peak to 1/e ∼37% of peak value). b, Decay constants of slow and fast cold tPN responses to a range of cold stimuli are systematically different (box-plot: red line, median; blue box, 25th and 75th percentiles; whiskers, data range; P < 0.001, t-test; ΔT = 2.91 ± 0.75, mean ± s.d.; n = 16 stimuli, n = 7 slow cells, 9 fast cells). In c, a larger cooling stimulus and the response it elicited in a representative slow-adapting and fast-adapting cell are shown side-by-side to facilitate comparison. Note that the slow-adapting cell displays a persistent response that lasts as long as the cooling stimulus. In contrast, as a consequence of faster decay, the response of a ‘fast-adapting’ cell returns to the pre-stimulus baseline when the temperature is still changing.
Extended Data Figure 5 ON and OFF responses amidst a complex temperature stimulus.
‘Fast-adapting cold’ (purple trace) and ‘fast-adapting hot’ cells (green trace) track remarkably well a dynamic temperature transient, displaying alternating ON and OFF responses. The cold cell type (purple trace) displays a significantly delayed response whenever a cooling phase is preceded by heating. Notably, cooling responses following a heating step begin when the temperature has nearly returned to baseline (red arrowheads referred to as OFF responses; see main text). Shown here is a representative single stimulus and corresponding response traces simultaneously recorded from adjacent cell bodies; genotype R95C02/UAS-GCaMP6m.
Extended Data Figure 6 ON and OFF responses of fast tPNs are not limited to a defined range of absolute temperature.
To better understand the nature of ON and OFF responses, we tested whether the dynamic activity of fast-adapting cells was limited to a defined range of absolute temperatures by delivering defined heating and cooling stimuli from a range of baseline temperatures. a, ON responses from cold (purple traces) and hot (green traces) cells recorded from a 25 °C baseline and from a higher (29 °C) or lower (20 °C) baseline, respectively (shown are representative single stimuli and corresponding response traces recorded from cell bodies). b, c, The activity of fast tPNs is not restricted to a specific temperature band as evident by plotting the absolute temperature corresponding to response onset (see arrows in a) against the baseline temperature for fast-adapting cold (b) and fast-adapting hot (c) cells, challenged with a range of hot and cold stimuli (ΔT 2–7° C; cooling ΔT = −4.7 ± 1.9 °C, heating ΔT = 4.6 ± 2 °C, mean ± s.d.; n > 10 cells/>10 animals per cell type). In b, c, filled purple and green dots represent ON responses while empty circles represent OFF responses. Owing to their specific delay, the OFF responses of cold cells start at the very end of a hot stimulus (that is, when the temperature has nearly returned to baseline Tonset – Tbaseline = 0.68 ± 0.43 °C, mean ± s.d.); in contrast, hot cells respond at or near the start of the heating phase that follows a cold stimulus (see below) and, as a result, systematically at a lower temperature than baseline (Tonset – Tbaseline = −2.97 ± 1.15 °C, mean ± s.d.; note that this value is significantly different than the one calculated for cold cells, P < 0.001, t-test). d–h, Fast-adapting hot cells innervating the lateral pathway display OFF responses whose delay varies by cell. d, e, A pair of adjacent hot-activated (green arrow and trace) and cold-activated (purple arrow and trace) tPNs innervating the lateral pathway (3D reconstruction in d, cell bodies in e; same cells and colour coding as in main Fig. 3). f, The cold-activated tPN responded to cooling stimuli with a rapid ON transient. The hot-activated cell in this pair displayed a calcium transient that was not delayed; that is, started immediately with the heating phase that followed the cooling stimulus (light green trace). g, h, In contrast, a different fast hot cell type in the same cluster (dark green circle and trace) displayed a delayed response (arrowhead), similar to the delayed OFF response reported for cold-activated tPNs at the end of a heating stimulus (see Fig. 3). Scale bars in e and g, 5 μm.
Extended Data Figure 7 Cold-specific tPNs innervating the mALT.
a–d, A distinct group of cold-activated tPNs follows the mALT and innervates the mushroom body and lateral horn (represented by a 3D reconstruction). b, The driver VT26020 is active in three neurons innervating this pathway (also showing prominent expression in auditory centres, AU, see Methods for details; PAL, posterior antennal lobe, the approximate position of the cold glomerulus is circled; genotype = VT26020-Gal4/UAS-CD8:GFP). c, Representative calcium-imaging traces from a single cell body displaying robust transients in response to cooling stimuli of increasing magnitude (note that these cells do not show activation in response to heating). d, Stimulus–response plot showing the responses of this cell type to defined cooling stimuli recorded at the cell body. The responses are proportional to cold stimuli over a wide range of intensities (R2 = 0.8, P < 0.001; slope = −0.45, 95% CI = [−0.57 to −0.33]; regression and slope statistics were calculated on all data points across individual flies). Each blue dot represents the maximal response of a single cell to a defined stimulus; dots are connected when the responses belong to the same cell, n = 3 cells/3 animals. Grey dots represent binned averages ± s.e.m. (bins = 2 °C; genotype: VT26020-Gal4/UAS-GCaMP6m). Scale bar in b, 25 μm.
Extended Data Figure 8 tPN pathways mediating temperature representation in the fly brain.
Summary of the array of tPN pathways mediating temperature representation in the fly brain, their properties and target regions. Multiple pathways carry specific information about stimulus quality (hot and cold) to the mushroom body, lateral horn and PLP. Most tPNs innervating these pathways are narrowly tuned, responding to either hot or cold stimuli, but a prominent group of cells is capable of responding to both heating and cooling temperature stimuli (broadly tuned). Moreover, response dynamics of tPNs are strikingly different, allowing a classification of tPN responses in slow versus fast adapting.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables listing the genotypes relevant to this study. (PDF 189 kb)
Rights and permissions
About this article
Cite this article
Frank, D., Jouandet, G., Kearney, P. et al. Temperature representation in the Drosophila brain. Nature 519, 358–361 (2015). https://doi.org/10.1038/nature14284
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14284
This article is cited by
-
Rapid threat assessment in the Drosophila thermosensory system
Nature Communications (2023)
-
Heterogeneous receptor expression underlies non-uniform peptidergic modulation of olfaction in Drosophila
Nature Communications (2023)
-
The cellular coding of temperature in the mammalian cortex
Nature (2023)
-
Integration of photoperiodic and temperature cues by the circadian clock to regulate insect seasonal adaptations
Journal of Comparative Physiology A (2023)
-
Organization of the parallel antennal-lobe tracts in the moth
Journal of Comparative Physiology A (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.