Abstract
The basal ganglia are phylogenetically conserved subcortical nuclei necessary for coordinated motor action and reward learning1. Current models postulate that the basal ganglia modulate cerebral cortex indirectly via an inhibitory output to thalamus, bidirectionally controlled by direct- and indirect-pathway striatal projection neurons (dSPNs and iSPNs, respectively)2,3,4. The basal ganglia thalamic output sculpts cortical activity by interacting with signals from sensory and motor systems5. Here we describe a direct projection from the globus pallidus externus (GP), a central nucleus of the basal ganglia, to frontal regions of the cerebral cortex (FC). Two cell types make up the GP–FC projection, distinguished by their electrophysiological properties, cortical projections and expression of choline acetyltransferase (ChAT), a synthetic enzyme for the neurotransmitter acetylcholine (ACh). Despite these differences, ChAT+ cells, which have been historically identified as an extension of the nucleus basalis, as well as ChAT− cells, release the inhibitory neurotransmitter GABA (γ-aminobutyric acid) and are inhibited by iSPNs and dSPNs of dorsal striatum. Thus, GP–FC cells comprise a direct GABAergic/cholinergic projection under the control of striatum that activates frontal cortex in vivo. Furthermore, iSPN inhibition of GP–FC cells is sensitive to dopamine 2 receptor signalling, revealing a pathway by which drugs that target dopamine receptors for the treatment of neuropsychiatric disorders can act in the basal ganglia to modulate frontal cortices.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
13 March 2015
Minor changes were made to Extended Data Figures 4, 6 and 8.
References
Yin, H. H. & Knowlton, B. J. The role of the basal ganglia in habit formation. Nature Rev. Neurosci. 7, 464–476 (2006)
Smith, Y., Bevan, M. D., Shink, E. & Bolam, J. P. Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience 86, 353–387 (1998)
Kravitz, A. V. et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature 466, 622–626 (2010)
Freeze, B. S., Kravitz, A. V., Hammack, N., Berke, J. D. & Kreitzer, A. C. Control of basal ganglia output by direct and indirect pathway projection neurons. J. Neurosci. 33, 18531–18539 (2013)
Goldberg, J. H., Farries, M. A. & Fee, M. S. Basal ganglia output to the thalamus: still a paradox. Trends Neurosci. 36, 695–705 (2013)
Seeman, P. Dopamine receptors and the dopamine hypothesis of schizophrenia. Synapse 1, 133–152 (1987)
Tohen, M. & Vieta, E. Antipsychotic agents in the treatment of bipolar mania. Bipolar Disord. 11, 45–54 (2009)
Bloch, M. H. et al. A systematic review: antipsychotic augmentation with treatment refractory obsessive-compulsive disorder. Mol. Psychiatry 11, 622–632 (2006)
Kita, H. Globus pallidus external segment. Prog. Brain Res. 160, 111–133 (2007)
Mesulam, M. M., Mufson, E. J., Levey, A. I. & Wainer, B. H. Atlas of cholinergic neurons in the forebrain and upper brainstem of the macaque based on monoclonal choline acetyltransferase immunohistochemistry and acetylcholinesterase histochemistry. Neuroscience 12, 669–686 (1984)
McKinney, M., Coyle, J. T. & Hedreen, J. C. Topographic analysis of the innervation of the rat neocortex and hippocampus by the basal forebrain cholinergic system. J. Comp. Neurol. 217, 103–121 (1983)
Grove, E. A., Domesick, V. B. & Nauta, W. J. Light microscopic evidence of striatal input to intrapallidal neurons of cholinergic cell group Ch4 in the rat: a study employing the anterograde tracer Phaseolus vulgaris leucoagglutinin (PHA-L). Brain Res. 367, 379–384 (1986)
Henderson, Z. The projection from the striatum to the nucleus basalis in the rat: an electron microscopic study. Neuroscience 78, 943–955 (1997)
Chang, H. T., Penny, G. R. & Kitai, S. T. Enkephalinergic-cholinergic interaction in the rat globus pallidus: a pre-embedding double-labeling immunocytochemistry study. Brain Res. 426, 197–203 (1987)
DeLong, M. R. Activity of pallidal neurons during movement. J. Neurophysiol. 34, 414–427 (1971)
Schultz, W. Changes in behavior-related neuronal activity in the striatum during learning. Trends Neurosci. 26, 321–328 (2003)
Laplane, D. et al. Obsessive-compulsive and other behavioural changes with bilateral basal ganglia lesions. A neuropsychological, magnetic resonance imaging and positron tomography study. Brain 112, 699–725 (1989)
Tkatch, T., Baranauskas, G. & Surmeier, D. J. Basal forebrain neurons adjacent to the globus pallidus co-express GABAergic and cholinergic marker mRNAs. Neuroreport 9, 1935–1939 (1998)
Mallet, N. et al. Dichotomous organization of the external globus pallidus. Neuron 74, 1075–1086 (2012)
Sarter, M. & Bruno, J. P. The neglected constituent of the basal forebrain corticopetal projection system: GABAergic projections. Eur. J. Neurosci. 15, 1867–1873 (2002)
Saunders, A., Johnson, C. A. & Sabatini, B. L. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front. Neural Circuits 6, 47 (2012)
Shepherd, G. M. & Harris, K. M. Three-dimensional structure and composition of CA3→CA1 axons in rat hippocampal slices: implications for presynaptic connectivity and compartmentalization. J. Neurosci. 18, 8300–8310 (1998)
Nambu, A., Tokuno, H. & Takada, M. Functional significance of the cortico-subthalamo-pallidal ‘hyperdirect’ pathway. Neurosci. Res. 43, 111–117 (2002)
Albin, R. L., Young, A. B. & Penney, J. B. The functional anatomy of basal ganglia disorders. Trends Neurosci. 12, 366–375 (1989)
Simpson, E. H., Kellendonk, C. & Kandel, E. A possible role for the striatum in the pathogenesis of the cognitive symptoms of schizophrenia. Neuron 65, 585–596 (2010)
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014)
Insel, T. R. Rethinking schizophrenia. Nature 468, 187–193 (2010)
Blum, B. P. & Mann, J. J. The GABAergic system in schizophrenia. Int. J. Neuropsychopharmacol. 5, 159–179 (2002)
Scarr, E., Gibbons, A. S., Neo, J., Udawela, M. & Dean, B. Cholinergic connectivity: it’s implications for psychiatric disorders. Front. Cell. Neurosci. 7, 55 (2013)
Lewis, D. A., Hashimoto, T. & Volk, D. W. Cortical inhibitory neurons and schizophrenia. Nature Rev. Neurosci. 6, 312–324 (2005)
Rossi, J. et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 13, 195–204 (2011)
Vong, L. et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011)
Taniguchi, H. et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011)
Gong, S. et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci. 27, 9817–9823 (2007)
Gerfen, C. R., Paletzki, R. & Heintz, N. GENSAT BAC Cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80, 1368–1383 (2013)
Tallini, Y. N. et al. BAC transgenic mice express enhanced green fluorescent protein in central and peripheral cholinergic neurons. Physiol. Genomics 27, 391–397 (2006)
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neurosci. 13, 133–140 (2010)
Madisen, L. et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nature Neurosci. 15, 1–12 (2012)
Tamamaki, N. et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J. Comp. Neurol. 467, 60–79 (2003)
Saunders, A., Johnson, C. A. & Sabatini, B. L. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front. Neural Circuits 6, 47 (2012)
Mesulam, M. M., Mufson, E. J., Levey, A. I. & Wainer, B. H. Cholinergic innervation of cortex by the basal forebrain: cytochemistry and cortical connections of the septal area, diagonal band nuclei, nucleus basalis (substantia innominata), and hypothalamus in the rhesus monkey. J. Comp. Neurol. 214, 170–197 (1983)
Beurrier, C., Ben-Ari, Y. & Hammond, C. Preservation of the direct and indirect pathways in an in vitro preparation of the mouse basal ganglia. Neuroscience 140, 77–86 (2006)
Marx, M., Günter, R. H., Hucko, W., Radnikow, G. & Feldmeyer, D. Improved biocytin labeling and neuronal 3D reconstruction. Nature Protocols 7, 394–407 (2012)
Freeze, B. S., Kravitz, A. V., Hammack, N., Berke, J. D. & Kreitzer, A. C. Control of basal ganglia output by direct and indirect pathway projection neurons. J. Neurosci. 33, 18531–18539 (2013)
Eberly, D., Gardner, R., Morse, B., Pizer, S. & Scharlach, C. Ridges for image analysis. J. Math. Imaging Vis. 4, 353–373 (1994)
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8, 679–698 (1986)
Acknowledgements
The authors thank the Lowell laboratory at Beth Israel Deaconess Medical Center for the gift of the DIO-synaptophysin-mCherry and DIO-synaptophysin-GFP rAAVs, R. Pemberton for technical support and F. Krienen, N. Duggan, P. Kaeser and members of the Sabatini laboratory for helpful discussions. This work was supported by grants from the National Institutes of Health (F31 NS074842) to A.S., (F31-MH093026-01A1) to I.A.O., (P30 EY12196) to the Vision Core and NINDS P30 Core Center grant (#NS072030) to the Neural Imaging Center and Neurobiology Imaging Center in the Department of Neurobiology at Harvard Medical School and a NIH grant (R01 NS046579) to B.L.S.
Author information
Authors and Affiliations
Contributions
A.S., I.A.O. and B.L.S. designed the experiments. A.S. performed the anatomical and acute slice experiments, analysed the data and assisted all other parts of the study. I.A.O. performed the in vivo recordings and analysed data. C.A.J. assisted with immunohistochemistry experiments and mouse genotyping. V.K.B. performed rhesus macaque anatomical experiments. C.R.G. sliced and imaged mouse brains for 3D reconstructions. N.D.K. performed the sectioning, staining and imaging for array tomography. H.L.E. and T.X. assisted in the image analysis for axon detection in whole-brain reconstructions and array tomography analysis, respectively. A.S. and B.L.S. wrote the manuscript with contributions from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Anatomical and molecular properties of GP–FC cells and ChAT+ cells of the substantia innominata and ventral pallidum.
a–c, GP–FC cells project exclusively to ipsilateral cortex. a, Low-magnification horizontal section from a wild-type mouse injected bilaterally in FC with red (right hemisphere) and green (left hemisphere) retrobeads. DAPI (blue), nuclear stain. Boxed insets show location of GP in b. Green signal in the right hemisphere is due to bleed through from the red channel. b, High-magnification of left and right GP from the same brain as in a. Retrobead+ cells from ipsilateral (ipsi) and contralateral (contra) injections are highlighted with white circles. Dashed lines demarcate the approximate boundaries of the GP. c, Summary graph showing nearly all retrobead+ cells (n = 436 of 437, from four mice) resulted from injections in ipsilateral FC. d, e, FC retrobead labelling in Vgati-Cre;Rosa26lsl-zsGreen or Gad2i-Cre;Rosa26lsl-zsGreen mice followed by ChAT immunostaining (magenta) demonstrates that nearly all retrobead+ GP cells (red) are Vgati-Cre-positive or Gad2i-Cre-postive (green) while a subset of retrobead+ neurons are also ChAT+ (solid circles) and the remainder are ChAT− (dashed circles). Nearly all retrobead+ GP cells were Vgat+ (n = 159 of 159 cells, from three mice) or Gad2+ (n = 231 of 233 cells, from two mice), whereas 72% were ChAT+ (n = 215 of 300 cells, from five mice) and 28% ChAT− (n = 85 of 300 cells). NB, nucleus basalis. d. Top, low-magnification sagittal view of the GP. Bottom, a single confocal plane from stacks used to quantify marker co-localization in Vgati-Cre;Rosa26lsl-zsGreen mice. e, A single confocal plane from stacks used to quantify marker co-localization in Gad2i-Cre;Rosa26lsl-zsGreen mice. f, g, ChAT+ neurons of the substantia innominata (SI) and ventral pallidum (VP) also express Vgat and Gad2. GPm, medial globus pallidus (entopeduncular nucleus). f, Top, low-magnification ventral view of a sagittal section from a Vgati-Cre;Rosa26lsl-zsGreen mouse immunostained for ChAT. Bottom, high-magnification view of the substantia innominata and bordering ventral pallidum. g, High-magnification view of the substantia innominata/ventral pallidum in a Gad2i-Cre;Rosa26lsl-zsGreen mouse immunostained for ChAT. h–j, ChAT− GP–FC cells do not express parvalbumin (PV). h, Low-magnification view of sagittal section through the GP of a wild-type mouse injected with retrobeads (red) in FC and immunostained for ChAT (green) and parvalbumin (magenta). White circles highlight retrobead+ GP–FC cells. i, A single confocal plane showing retrobead+ GP–FC cells (circled in white) and immunostaining for ChAT and parvalbumin. j, Confocal quantification of co-localization between retrobead+ ChAT− GP–FC cells and parvalbumin (n = 32 cells, from two mice).
Extended Data Figure 2 ChAT+ and ChAT− GP–FC cells are present in a rhesus macaque.
a, In order to label frontal cortical projection neurons from Ch4iv and Ch4id regions of the nucleus basalis adjacent to the GP of a rhesus macaque41, the neuronal tracer biotinylated dextran amine (BDA) was injected at multiple sites along the arcuate and principal gyri and in the orbital cortex. Left, dorsal (top) and ventral (bottom) views of a fixed macaque brain. Dashed boxes indicate the injected areas. Right, schematic of the injection sites. Blue circles correspond to 2 × 0.5 μl BDA injections at 1 and 2 mm below the pial surface. b, Coronal section through the injection area after immunostaining to visualize BDA. c–e, Immunostaining for BDA and ChAT identifies retrogradely labelled ChAT+ and ChAT− GP–FC cells. c, Coronal section from macaque atlas containing GP and nucleus basalis. d, Left, ChAT immunostaining highlights traditional anatomical boundaries of the GP–putamen and GP–nucleus basalis. Same plane as in c. Right, higher magnification view of GP–nucleus basalis border corresponding to the inset in c. ChAT+ neurons are distributed around the ventral GP/dorsal nucleus basalis and along laminae separating the GP from the putamen (lateral medullary laminae, lml) and globus pallidus internus (medial medullary laminae, mml). Arrow and arrowhead indicate approximate locations of BDA+ChAT+ (680 μm anterior) and BDA+ChAT− (360 μm anterior) example GP–FC cells shown in e. e, Single confocal planes showing example BDA+ChAT+ and BDA+ChAT− GP–FC cells. f, g, ChAT immunostaining in a Drd2-eGFP mouse distinguishes traditional anatomical boundaries of the GP–nucleus basalis from the territory occupied by iSPN axons. f, Coronal section from the mouse atlas. g, Left, ChAT immunostaining highlights traditional anatomical boundaries of the GP–striatum and GP–nucleus basalis. Same plane as in f. Right, higher magnification view of GP–nucleus basalis border region corresponding to the inset in f. As in macaque, ChAT+ cells are distributed along GP borders between striatum and the internal capsule (ic) and at the border of ventral GP–dorsal nucleus basalis. Overlay of GFP fluorescence demonstrates iSPN axons arborize throughout the GP, abutting ChAT+ cells on the GP border regions (arrow), and ventrally in the dorsal nucleus basalis (arrowhead). AA, anterior amygdaloid area; ac, anterior commissure posterior; CeL, central lateral division of amygdala; CeM, central medial division of amygdala; GP, globus pallidus externus; GPi, globus pallidus internus; Lh, lateral hypothalamus; Pu, putamen; SI, substantia innominata; Str, striatum.
Extended Data Figure 3 Validation of ChATi-Cre knock-in mouse and rAAV strategy for Cre-On/Off labelling of GP–FC axons in cortex.
a–c, The ChATi-Cre mouse expresses Cre selectively in ChAT+ GP/nucleus basalis neurons with high penetrance. a, Left, low-magnification view of sagittal section through ChATi-Cre;Rosa26lsl-tdTomato mouse immunostained for ChAT. Right, inset showing higher magnification view of the GP and nucleus basalis. Dashed line approximates the boundaries for quantifying Cre-reporter and ChAT co-localization. b, Single confocal plane showing overlap of Cre-reporter expression and ChAT immunostaining of ChAT+ cells at the GP–nucleus basalis border. c, Quantification of confocal co-localization between the Cre-reporter+ and ChAT+ cells (n = 471 of 484 tdTomato+ ChAT+, n = 12 of 484 tdTomato+ only, n = 1 of 484 ChAT+ only, from three mice). d–f, Transduction of the GP in a ChATi-Cre mouse with DIO-eGFP (Cre-On) and FAS–tdTomato (Cre-Off) rAAVs effectively targets GFP and tdTomato to ChAT+ and ChAT− cells respectively. d, Sagittal section through the GP following transduction of the DIO-eGFP (green) and FAS–tdTomato (red) rAAVs and immunostaining for ChAT (magenta). e, Single confocal plane showing ChAT+ cells (circled in white) co-localized with GFP but not tdTomato. f, Quantification of confocal co-localization between ChAT, GFP and tdTomato (n = 319 ChAT+ cells, from two mice). g, Left, coronal atlas. Right, injection site for injection 1 (Fig. 1b–d), showing ChAT+ (Cre-On) expression limited to GP and the immediately adjacent nucleus basalis. ChAT− (Cre-Off) expression is limited to GP with slight leak in striatum. ChAT+ and ChAT− axons arborize in the thalamic reticular nucleus (Rt). SI, substantia innominata; NB, nucleus basalis; ic, internal capsule. h, Coronal section of anterior M1 illustrating automated axon detection of ChAT− and ChAT+ axons. i, Densities of detected ChAT− (green) and ChAT+ (magenta) axons along with total cortical area (black) in successive 50-μm coronal slices (anterior–posterior) across the cortical mantle (n = 2 mice). Densities and area are shown raw (thin line) and after five point median filtering (thick line). Median-filtered data are reported in Fig. 1c. j, Average density of ChAT+ and ChAT− GP–FC axons by cortical area. FrA, frontal association; PrL, prelimbic; MO, medial orbital; LO, lateral orbital; VO, ventral orbital; GI/DI, granular/dysgranular insular; AI, agranular insular; M1, primary motor; M2, secondary motor; S1, primary sensory; S2, secondary sensory; Cg, cingulate; IL, infralimbic; DLO, dorsolateral orbital; Pir, piriform; A2, secondary auditory; A1, primary auditory; RS retrosplenial; V1, primary visual; V2, secondary visual; TeA, temporal association; PTA, parietal; Ent, enthorhinal; Ect, ectorhinal; PRh, perirhinal. k, Coronal section illustrating the distribution of GP–FC axons across layers of ectorhinal cortex, a posterior cortical area that receives a large GP–FC projection. ChAT− axons target superficial layers 1 and 2/3 (arrow), in addition to deeper layers 5 and 6, as in anterior cortices including M1. The ChAT+ axons arborize across all cortical layers in both ectorhinal and anterior cortical areas. rh, rhinal fissure; Str, striatum; wm, white matter.
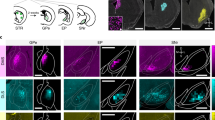
Extended Data Figure 4 ChAT+ and ChAT− GP–FC cells target distinct but overlapping subcortical nuclei.
a, Coronal sections from 3D brain reconstructions illustrating subcortical nuclei targeted by ChAT+ and ChAT− axons following transduction of ChATi-Cre GP/dorsal nucleus basalis with rAAVs DIO-eGFP (magenta, Cre-On) and FAS–tdTomato (green, Cre-Off) (n = 2 mice, examples from injection 1). Left, coronal atlas. Right, high-magnification views of subcortical nuclei.
Extended Data Figure 5 GP–FC cells are distinguished by active and passive membrane properties.
a, Maximum intensity two-photon projections of example ChAT− (left) and ChAT+ (right) GP–FC cells following whole-cell recording. Dashed insets show high-magnification projections through dendrites. b, ChAT+ cells have larger soma than ChAT− cells. Soma volumes were quantified from two-photon stacks (from four P18–22 mice; n = 8 ChAT+ cells, n = 8 ChAT− cells). c, Passive membrane properties of GP–FC cells (n = 9 ChAT−, n = 10 ChAT+, from four P18–22 mice). d, Representative waveforms of spontaneously active ChAT− and ChAT+ GP–FC cells. e, Schematic of quantified membrane properties following positive and negative current injections. ISI, interspike interval. f, Active membrane properties of GP–FC cells (n = 8–9 ChAT−, n = 10 ChAT+). Representative action potential waveforms from a spontaneously active ChAT− cell or from minimal current injection in a ChAT+ cell. Evoked firing rates were calculated for 500-ms current injections. P < 0.05 (Mann–Whitney test). g, Developmental comparison of GP–FC membrane properties before (P13–14, n = 2 mice) and around (P50–56, n = 3 mice) sexual maturity. Left, ChAT− GP–FC cells are spontaneously active throughout postnatal development (P13–14, n = 4 of 4; P50–56, n = 4 of 5), while ChAT+ cells tend to become spontaneously active after sexual maturity (P13–14, n = 1 of 11; P50–56, n = 7 of 8). GP–FC membrane resistance (middle) does not change before and after sexual maturity but membrane capacitance (right) is reduced (ChAT−: P13–14, n = 4; P50–56, n = 5; ChAT+: P13–14, n = 9; P50–56, n = 9). P < 0.05 (Fisher’s exact test); error bars denote mean ± s.e.m.
Extended Data Figure 6 Optogenetic manipulations of GP–FC cells coupled with in vivo extracellular FC recordings in an awake behaving mouse.
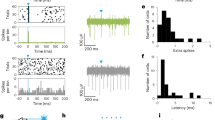
a, Summary plot of mean (and 95% confidence interval) indices for light modulation (Ilight) of FC unit firing rate by experimental condition. Dashed line denotes average of blue light fibre mCherry+ control. The number of mice for each condition are shown in parentheses. P < 0.0001, Kruskal–Wallis test. b, Pulsed ChR2 depolarization of Vgati-Cre GP somata increases firing rates in FC on a millisecond timescale. Left, experimental schematic. Extracellular recordings in FC are coupled with fibre-delivered pulses of blue light (5-ms pulses of 473 nm, 10 Hz for 3 s) in GP. Middle, mean firing rate (±s.e.m.) of all FC units in response to pulsed blue light in ChR2+ (blue) or control (black) mice (ChR2+, n = 90 units from five mice; mCherry+, n = 99 units from three mice). Dotted line represents mean pre-stimulation firing rate. Right, pseudo-coloured plot of normalized firing rate for all units. c, Fibre-delivered pulsed blue light illumination of mCherry+ GP somata in ChATi-Cre mice shows no light-induced changes in the firing rates of FC units above chance (increased, 0 of 99; decreased, 2 of 99; from three mice). Right, pseudo-coloured plot of normalized firing rate for all units. d, Fibre-delivered constant yellow light illumination of mCherry+ GP somata in ChATi-Cre mice shows no light-induced changes in the firing rates of FC units above chance (increased, 1 of 63; decreased, 2 of 63; from two mice;). Right, pseudo-coloured plot of normalized firing rate for all units. e, Latencies of light-induced modulation of FC firing following fibre-based illumination of ChR2+ (pulsed blue light) or ArchT+ (constant yellow light) GP somata in Vgati-Cre mice. Top, spike raster plots (upper) and firing rate histograms (lower, 50-ms bins) from example FC units exhibiting light-induced decreases and increases in firing rates. Onset times for light pulses are shown with coloured dash lines. Bottom, summary graph plotting light-effect latencies for those FC units with statistically significant modulations. Firing rates are binned every 50 ms, such that ‘Bin of first change' = 0 contains the spikes from 0–50 ms after light onset. First change is determined as the first bin to deviate more than ±2 s.d. from the mean baseline firing. Units in which no change is detected within 500 ms are excluded. Individual units may have a first increasing and first decreasing bin if their activity is biphasic. Mean 50 ms bin of first activated by ArchT in Vgati-Cre mice was 3.0 ± 0.5, n = 47 of 96 units; first suppressed 4.3 ± 0.5, n = 28 of 96 units; first activated by ChR2 2.9 ± 0.3, n = 22 of 90 units, first suppressed by ChR2 0.7 ± 0.2, n = 35 of 90 units. f, Optrode-delivered pulsed blue light illumination of ChR2+ axons in FC from Vgati-Cre mice shows no persistent changes in the firing rates of FC units above chance (increased, 1 of 111; decreased, 2 of 111 from five mice). Right, pseudo-coloured plot of normalized firing rate for all units. g, Pulsed blue light illumination of GP–FC axons using an optrode leads to increases in firing rate on the millisecond timescale. Left, mean (±s.e.m.) firing rates on a millisecond time scale of all units in response to pulsed blue light illumination of GP axons in FC for Vgati-Cre mice expressing ChR2 (blue) or mCherry (black) or ChATi-Cre mice expressing ChR2 (purple). Right, z score of inter-pulse interval firing rates (20 × 5-ms bins) comparing positive and negative deviations from preceding baseline period without light. h, Optrode-delivered pulsed blue light illumination of ChR2+ axons in FC from ChATi-Cre mice shows no persistent changes in the firing rates of FC units above chance (increased, 0 of 74 from three mice; decreased, 0 of 74). Right, pseudo-coloured plot of normalized firing rate for all units. i, Fibre-delivered constant yellow light illumination of ArchT+ GP somata in ChATi-Cre mice shows no light-induced changes in the firing rates of FC units above chance (increased, 1 of 120; decreased, 3 of 120 from four mice). Right, pseudo-coloured plot of normalized firing rate for all units. In plots of Ilight, red bars indicate units that were statistically significantly modulated by light (t-test, P < 0.05). For pseudo-coloured plots of normalized firing rate, units are normalized to the baseline period, before light onset, and ordered by Ilight (low to high). Blues and purples represent low firing rates whereas yellow and red represent higher firing rates. Red represents modulations three or more times baseline.
Extended Data Figure 7 ChR2-mediated stimulation of ChATi-Cre axons following rAAV transduction or with a Cre-activated allele evokes ACh- and GABA-mediated currents.
a, b, Targeting ChR2 expression to ChAT+ and ChAT− GP–FC cells. a, Schematic of ChAT+ (magenta) or ChAT− (black) GP–FC axons expressing ChR2–mCherry after DIO (Cre-On) or DO (Cre-Off) rAAV transduction in the GP of ChATi-Cre;GAD1GFP mice. b, rAAV DO-ChR2-mCherry transduced into the ChATi-Cre GP expresses ChR2-mCherry selectively in Cre− neurons. Single confocal plane showing neighbouring ChR2-mCherry+ soma (dotted outline) and ChAT+ soma at the GP–nucleus basalis border. Of 158 ChR2-mCherry+ neurons surrounding 223 ChAT+ neurons, 0 were ChR2-mCherry+ChAT+ (from two mice). c, ChAT+ axons surrounding GAD1GFP expressing cells in FC layer 6. d–f, ChAT+ GP–FC cells ramify local axon collaterals around the GP–nucleus basalis border. d, Sagittal atlas with the GP–nucleus basalis border indicated with a dashed box. e, Left, low-magnification view of ChATi-Cre GP following transduction with rAAV DIO-synaptophysin-mCherry. DAPI (grey), nuclear stain. Right, maximum projection confocal stack (28 μm) of inset region. Example putative presynaptic boutons indicated by arrowheads. f, Left, low-magnification sagittal section from ChATi-Cre;Rosa26lsl-ChR2-eYFP/+ mouse. Right, high-magnification inset of GP showing distribution of neurons (NeuN immunostain, magenta) and ChR2-eYFP+ processes (white). g, h, Following rAAV transduction in ChATi-Cre mice, ChR2 activation of local ChATi-Cre axon collaterals results in rare nicotinic EPSCs but prevalent GABAergic IPSCs onto ChR2− GP/nucleus basalis neurons (EPSC = 2 of 85 cells; IPSC = 7 of 85 cells from six mice). g, Light-evoked EPSC from an example ChR2− GP/nucleus basalis cell voltage-clamped at −70 mV (top) was insensitive to glutamate receptor block with NBQX and CPP (middle), but abolished by bath application of MEC, MLA & DHßE (bottom), suggesting the EPSC resulted from ACh release and nicotinic receptor activation. h, Summary of peaks from nicotinic EPSCs and GABAergic IPSCs evoked from ChATi-Cre axons onto ChR2− GP/nucleus basalis cells. i, Left, low-magnification image of sagittal section from a ChATi-Cre;Rosa26lsl-ChR2-eYFP/+ mouse. Right, high-magnification of inset from frontal cortex showing distribution of neurons (NeuN immunostain, magenta) and ChR2-eYFP (white), expressed in axons from basal forebrain and in local cortical interneurons. j, Maximum intensity two-photon projection of a layer 1 interneuron following whole-cell recording. k, Left, light-evoked current responses from two layer 1 interneurons held at indicated potentials to optogenetic activation in a ChATi-Cre;Rosa26lsl-ChR2-eYFP/+ mouse in baseline conditions (black, NBQX & CPP) and after bath application of nicotinic receptor antagonist cocktail (red, MEC, MLA & DHßE). Right, nicotinic EPSCs are blocked by bath application of the non-selective blocker MEC alone (green). l, Time until full block of light-evoked nicotinic EPSCs following bath application of either nicotinic receptor antagonist cocktail (MEC, MLA & DHβE, n = 5 cells from three mice) or MEC alone (n = 5 cells from three mice). Inter-stimulus interval = 20 s.
Extended Data Figure 8 Synaptic connectivity and array tomography marker co-localization analysis of GP–FC axons in FC.
a–d, Ionotropic synaptic connectivity of ChAT+ and ChAT− GP–FC neurons onto FC cell types and layers. a, Example morphologies of FC neurons identified as pyramidal (from layer 5) or an interneuron (from layer 1). b, Summary of cortical neurons with ChR2-evoked monosynaptic ionotropic GABAergic or nicotinic currents from ChAT+ or ChAT− axons by cortical layer. c, Peak currents induced by ChR2 activation of ChAT+ or ChAT− GP–FC cells in FC. Postsynaptic cells are grouped across layers as pyramidal or interneurons. Left, GABAergic IPSCs reported with either TTX/4AP in the bath or following wash-in are plotted with dotted data. IPSCs recorded in baseline conditions (ChAT−, NBQX & CPP; ChAT+, ACSF only) are plotted with undotted data. Each cell is represented once. (ChAT−: n = 5 pyramidal, n = 15 interneurons from 13 mice; ChAT+: n = 3 interneurons from 15 mice). Right, nicotinic EPSCs recorded in ACSF, present after bath application of CPP and NBQX and fully blocked by nicotinic receptor antagonists (MEC, MLA, DHßE, n = 5 interneurons from 15 mice). d, Onset latencies for IPSCs and EPSCs induced by ChR2 activation of ChAT+ or ChAT− GP–FC cells under baseline conditions only (ACSF) or in the presence of TTX/4AP. ‘BaselineTTX/4AP’ refers to the subset of cells with IPSCs recorded under both baseline conditions and recovered following wash-in of TTX/4AP (n = 5; same data as Fig. 3b). Onset latencies of ChAT− IPSCs were longer in TTX/4AP (n = 14) than ACSF (n = 11). P < 0.05 (Mann–Whitney); error bars denote mean ± s.e.m. e–k, Array-tomography-based co-localization analysis of ChAT+ presynaptic terminals (PSTs) in FC. e, Left, 300 μm sagittal slab from a ChATi-Cre mouse injected with 300 nl of rAAV DIO-synaptophysin-GFP in GP. Right, box indicates area of FC prepared for array tomography. f, Automated detection of GFP+ volumes (pearls) and synaptic markers. Left, maximum projection of GFP+ axons following computational detection of string-associated pearls (in red). Right, a single 70 nm plane showing diffraction-limited immunohistochemical punctae for PSD-95 and computational detection of point sources (in red). g, Maximum projection (z = 2.17 μm) through layers 1–3 of FC following injection of rAAV DIO-synaptophysin-GFP into the GP of a ChATi-Cre mouse. Inset shows location of axon shown in Fig. 3c. h–k, GFP+ pearls are putative GABAergic PSTs. h, Individual volumes for all detected GFP+ PSTs (n = 6,071 pearls from two mice; n = 4 layers 1 & 2/3 stacks, n = 4 layer 5 stacks). i, Mean density by distance plots for GFP+ PSTs versus synapsin I, bassoon, GAD1/2, VAChT, VGAT, gephyrin, PSD-95 and parvalbumin from example stack. Crosses indicate means from real data, while lines denote mean values following 1,000 rounds of marker randomization. Error bars denote 99% confidence intervals. j, Mean densities within GFP+ PSTs (0 distance) for all markers and all stacks. Real and randomized data are indicated as in i. k, z score summary (n = 8 stacks) quantifying the differences in mean density for the synaptic markers shown in j (and not reported in Fig. 3e) within GFP+ PSTs (0 distance) for the real data and following ten rounds of randomization of GFP+ volumes. Positive z scores indicate higher densities in the real data. P < 0.001 for all stacks.
Extended Data Figure 9 GP–FC cells receive glutamatergic synapses from the STN.
a, Left, low-magnification view of parasagittal slice showing the GP following biocytin labelling of the STN and avidin-HRP/DAB visualization of STN projections. Right, high-magnification view of inset showing DAB-labelled projections in the GP and around the GP–nucleus basalis border. b, Schematic of experimental strategy to electrically stimulate STN projections to GP. A bipolar electrode was placed at the anterior border of STN and GP–FC cells were targeted for whole-cell voltage-clamp recording. c, Acute parasagittal slice showing location of the bipolar electrode and recording pipette (red asterisk). d, e, Electrically evoked glutamatergic currents in GP–FC cells following stimulation of STN–GP axon tract. d, Left, example NBQX-subtracted AMPAR (Vhold = −70 mV) then CPP-subtracted NMDAR (Vhold = +40 mV) currents evoked in GP–FC cells under baseline conditions (SR95331, scopolamine, CGP55845). Right, summary of AMPAR and NMDAR peak currents in ChAT+ (n = 6) and ChAT− (n = 4) GP–FC cells (from seven mice). e, Onset latencies of glutamatergic currents (ChAT+: 2.3 ± 0.3 ms, n = 6 cells; ChAT−: 3.08 ± 0.3 ms, n = 4 cells, from seven mice). Bars denote mean ± s.e.m.
Extended Data Figure 10 GP–FC cells receive GABAergic synapses from dorsal striatal iSPNs and dSPNs with different presynaptic properties.
a, Left, sagittal sections from an Adora2a-Cre;Rosa26lsl-ChR2-eYFP/+ (top) or Drd1a-Cre;Rosa26lsl-ChR2-eYFP/+ (bottom) mouse where ChR2–eYFP is expressed in either iSPNs or dSPNs, respectively. Right, light was delivered first over the recorded cell (peak IPSCs in b) and then in dorsal striatum (pharmacological analysis in Fig. 4c and Extended Data Fig. 10h and presynaptic properties in f, g). c, d, SPNs from dorsal striatum arborize axons in and around the GP–nucleus basalis border but not in the basal forebrain. c, Left, sagittal section from a VGATi-Cre mouse injected with rAAV DIO-mCherry into dorsal striatum. Right, higher-magnification view of inset. Axons from SPNs arborize in the GP proper and GP–nucleus basalis border regions (areas 1 and 2) but not in the more ventral region of the basal forebrain (area 3, nucleus basalis proper/substantia innominata). d, Left, example maximum projection confocal stacks (z = 28–42 μm) from the inset regions in c. Right, binary axonal quantification from the regions indicated. SPN axon density is sharply reduced in nucleus basalis proper/substantia innominata. e, Synaptic connectivity screen for dorsal striatal SPN IPSCs onto ChAT+ neurons of the GP and basal forebrain. ChAT–GFP mice were injected with Cre-Off rAAV DO-ChR2-mCherry in dorsal striatum and whole-cell recordings were targeted to ChAT+ neurons (n = 23 cells, from four mice) in combination with optogenetic activation. NBQX and CPP were included in the bath to block glutamatergic transmission. Left, sagittal map of recording locations of ChAT−GFP+ neurons with detected IPSCs (blue) and no detected IPSCs (red). Right, peaks and onset latencies for detected IPSCs. Green lines denote means. In every experiment, IPSCs onto ChAT−GFP+ neurons of the GP were detected before recordings were targeted to more ventral areas. f, g, Paired pulse optogenetic activation of iSPNs (Adora2a-Cre;Rosa26lsl-ChR2-eYFP/+) and dSPNs (Drd1a-Cre;Rosa26lsl-ChR2-eYFP/+) in dorsal striatum reveals differences in short-term synaptic plasticity properties in GP–FC cells. f, Examples of optogenetically evoked paired pulse IPSCs (interstimulus interval = 20, 50, 100, 200 and 500 ms) from iSPNs (left) and dSPNs (right) in GP–FC cells. g, Mean 2nd/1st peak IPSC ratios (iSPN: n = 13 ChAT+, n = 9 ChAT− cells from 11 mice; dSPN: n = 9 ChAT+, n = 8 ChAT− cells from six mice). P < 0.05 iSPNs versus dSPNs (ChAT+ and ChAT− grouped together, Mann–Whitney); error bars denote s.e.m. h, Example iSPN peak IPSCs in a ChAT− GP–FC cell following application of quinpirole, sulpiride and SR95331. Inset, average IPSCs. Scale bar, 20 pA/10 ms. Rs, series resistance.
Supplementary information
Three-dimensional whole-brain reconstructions of axonal projections from ChAT+ (magenta) and ChAT- (green) GP-FC cells.
rAAVs DIO-EGFP (Cre-On) and FAS-tdTomato (Cre-Off) were injected into the GP and adjacent dorsal NB in a ChAT i-Cre/+ mouse. The max projection video begins with a lateral to medial view of the injection site and Rt. A posterior to anterior fly through of cortex shows axons in frontal cortex and the lateral amygdala, followed by a medial to lateral view that exhibits subcortical projections to the Str, STN, SNr/c, PF and LH. A small amount of FAS-tdTomato expression in striatum contributes to the Cre-Off projection in SNr. Rt, thalamic reticular nucleus; Str, striatum; SNr/c, substantia nigra reticulata/compacta; PF, thalamic parafascicular nucleus; LH, lateral habenula. (MOV 28915 kb)
Three-dimensional whole-brain reconstructions of axonal projections from iSPNs (green) and dSPNs (magenta) from dorsal striatum into GP/dorsal NB.
rAAVs DIO-EGFP (Cre-On) and FAS-tdTomato (Cre-Off) were injected into the dorsal striatum of D2r-Cre mouse, to differentially label Cre+ iSPNs and Cre- non-iSPNs. Since SPNs provide the only output of striatum, this strategy selectively labels iSPN and dSPN projections. Following a rotated lateral view of the whole brain, the video proceeds anterior to posterior, following single coronal slices through the injection site and back and forth through the GP before continuing to the SNr. Visualizing iSPNs and dSPNs individually illustrates that dSPN axons arborize widely in the ventral and posterior regions of GP/NB containing GP-FC cells. (MOV 17684 kb)
Rights and permissions
About this article
Cite this article
Saunders, A., Oldenburg, I., Berezovskii, V. et al. A direct GABAergic output from the basal ganglia to frontal cortex. Nature 521, 85–89 (2015). https://doi.org/10.1038/nature14179
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14179
This article is cited by
-
Updating the striatal–pallidal wiring diagram
Nature Neuroscience (2024)
-
A sleep-active basalocortical pathway crucial for generation and maintenance of chronic pain
Nature Neuroscience (2023)
-
Cell and circuit complexity of the external globus pallidus
Nature Neuroscience (2023)
-
A non-canonical striatopallidal Go pathway that supports motor control
Nature Communications (2023)
-
Mathematical derivation and mechanism analysis of beta oscillations in a cortex-pallidum model
Cognitive Neurodynamics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.