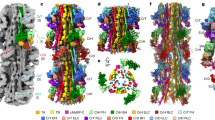
Abstract
Filamentous actin (F-actin) is the major protein of muscle thin filaments, and actin microfilaments are the main component of the eukaryotic cytoskeleton. Mutations in different actin isoforms lead to early-onset autosomal dominant non-syndromic hearing loss1, familial thoracic aortic aneurysms and dissections2, and multiple variations of myopathies3. In striated muscle fibres, the binding of myosin motors to actin filaments is mainly regulated by tropomyosin and troponin4,5. Tropomyosin also binds to F-actin in smooth muscle and in non-muscle cells and stabilizes and regulates the filaments there in the absence of troponin6. Although crystal structures for monomeric actin (G-actin) are available7, a high-resolution structure of F-actin is still missing, hampering our understanding of how disease-causing mutations affect the function of thin muscle filaments and microfilaments. Here we report the three-dimensional structure of F-actin at a resolution of 3.7 Å in complex with tropomyosin at a resolution of 6.5 Å, determined by electron cryomicroscopy. The structure reveals that the D-loop is ordered and acts as a central region for hydrophobic and electrostatic interactions that stabilize the F-actin filament. We clearly identify map density corresponding to ADP and Mg2+ and explain the possible effect of prominent disease-causing mutants. A comparison of F-actin with G-actin reveals the conformational changes during filament formation and identifies the D-loop as their key mediator. We also confirm that negatively charged tropomyosin interacts with a positively charged groove on F-actin. Comparison of the position of tropomyosin in F-actin–tropomyosin with its position in our previously determined F-actin–tropomyosin–myosin structure8 reveals a myosin-induced transition of tropomyosin. Our results allow us to understand the role of individual mutations in the genesis of actin- and tropomyosin-related diseases and will serve as a strong foundation for the targeted development of drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The coordinates for the electron microscope structure has been deposited in the Electron Microscopy Data Bank under accession code EMD-6124. Coordinates of F-actin in complex with tropomyosin have been deposited in the Protein Data Bank with accession number 3J8A.
References
van Wijk, E. et al. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26). J. Med. Genet. 40, 879–884 (2003)
Milewicz, D. M. et al. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu. Rev. Genomics Hum. Genet. 9, 283–302 (2008)
Sparrow, J. C. et al. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul. Disord. 13, 519–531 (2003)
Gordon, A. M., Homsher, E. & Regnier, M. Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 (2000)
Maytum, R., Lehrer, S. S. & Geeves, M. A. Cooperativity and switching within the three-state model of muscle regulation. Biochemistry 38, 1102–1110 (1999)
Pittenger, M. F., Kazzaz, J. A. & Helfman, D. M. Functional properties of non-muscle tropomyosin isoforms. Curr. Opin. Cell Biol. 6, 96–104 (1994)
Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F. & Holmes, K. C. Atomic structure of the actin:DNase I complex. Nature 347, 37–44 (1990)
Behrmann, E. et al. Structure of the rigor actin-tropomyosin-myosin complex. Cell 150, 327–338 (2012)
Holmes, K. C., Popp, D., Gebhard, W. & Kabsch, W. Atomic model of the actin filament. Nature 347, 44–49 (1990)
Fujii, T., Iwane, A. H., Yanagida, T. & Namba, K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467, 724–728 (2010)
Murakami, K. et al. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell 143, 275–287 (2010)
Galkin, V. E., Orlova, A., Schröder, G. F. & Egelman, E. H. Structural polymorphism in F-actin. Nature Struct. Mol. Biol. 17, 1318–1323 (2010)
Sousa, D. R., Stagg, S. M. & Stroupe, M. E. Cryo-EM structures of the actin:tropomyosin filament reveal the mechanism for the transition from C- to M-state. J. Mol. Biol. 425, 4544–4555 (2013)
Oda, T., Iwasa, M., Aihara, T., Maéda, Y. & Narita, A. The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009)
Laing, N. G. et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1). Hum. Mutat. 30, 1267–1277 (2009)
Oztug Durer, Z. A., Diraviyam, K., Sept, D., Kudryashov, D. S. & Reisler, E. F-actin structure destabilization and DNase I binding loop fluctuations: mutational cross-linking and electron microscopy analysis of loop states and effects on F-actin. J. Mol. Biol. 395, 544–557 (2010)
Strzelecka-Gołaszewska, H., Mossakowska, M., Woźniak, A., Moraczewska, J. & Nakayama, H. Long-range conformational effects of proteolytic removal of the last three residues of actin. Biochem. J. 307, 527–534 (1995)
Crosbie, R. H. et al. Structural connectivity in actin: effect of C-terminal modifications on the properties of actin. Biophys. J. 67, 1957–1964 (1994)
Hung, R.-J. et al. Mical links semaphorins to F-actin disassembly. Nature 463, 823–827 (2010)
Hung, R.-J., Pak, C. W. & Terman, J. R. Direct redox regulation of F-actin assembly and disassembly by Mical. Science 334, 1710–1713 (2011)
Iwasa, M., Maeda, K., Narita, A., Maéda, Y. & Oda, T. Dual roles of Gln137 of actin revealed by recombinant human cardiac muscle alpha-actin mutants. J. Biol. Chem. 283, 21045–21053 (2008)
Vorobiev, S. et al. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl Acad. Sci. USA 100, 5760–5765 (2003)
Koy, A. et al. Nemaline myopathy with exclusively intranuclear rods and a novel mutation in ACTA1 (Q139H). Neuropediatrics 38, 282–286 (2007)
Li, X. E. et al. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys. J. 100, 1005–1013 (2011)
Lorenz, M., Poole, K. J., Popp, D., Rosenbaum, G. & Holmes, K. C. An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin-tropomyosin gels. J. Mol. Biol. 246, 108–119 (1995)
Lehman, W., Orzechowski, M., Li, X. E., Fischer, S. & Raunser, S. Gestalt-binding of tropomyosin on actin during thin filament activation. J. Muscle Res. Cell Motil. 34, 155–163 (2013)
Poole, K. J. V. et al. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 155, 273–284 (2006)
Holmes, K. C. & Lehman, W. Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 29, 213–219 (2008)
Pardee, J. D. & Spudich, J. A. Purification of muscle actin. Methods Enzymol. 85B, 164–181 (1982)
Coulton, A., Lehrer, S. S. & Geeves, M. A. Functional homodimers and heterodimers of recombinant smooth muscle tropomyosin. Biochemistry 45, 12853–12858 (2006)
Hohn, M. et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Dominguez, R. & Holmes, K. C. Actin structure and function. Annu. Rev. Biophys. 40, 169–186 (2011)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Penczek, P. A. et al. CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy 140, 9–19 (2014)
Penczek, P. A. Resolution measures in molecular electron microscopy. Methods Enzymol. 482, 73–100 (2010)
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010)
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993)
Wang, Z. & Schröder, G. F. Real-space refinement with DireX: from global fitting to side-chain improvements. Biopolymers 97, 687–697 (2012)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010)
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010)
Li, X. E., Orzechowski, M., Lehman, W. & Fischer, S. Structure and flexibility of the tropomyosin overlap junction. Biochem. Biophys. Res. Commun. 446, 304–308 (2014)
Lehman, W., Li, X. E., Orzechowski, M. & Fischer, S. The structural dynamics of α-tropomyosin on F-actin shape the overlap complex between adjacent tropomyosin molecules. Arch. Biochem. Biophys. 552–553, 68–73 (2014)
Anandakrishnan, R., Aguilar, B. & Onufriev, A. V. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541 (2012)
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005)
Stenson, P. D. et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133, 1–9 (2014)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Hayward, S. & Lee, R. A. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J. Mol. Graph. Model. 21, 181–183 (2002)
Otterbein, L. R., Graceffa, P. & Dominguez, R. The crystal structure of uncomplexed actin in the ADP state. Science 293, 708–711 (2001)
Bathe, F. S., Rommelaere, H. & Machesky, L. M. Phenotypes of myopathy-related actin mutants in differentiated C2C12 myotubes. BMC Cell Biol. 8, 2 (2007)
Costa, C. F. et al. Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J. Cell Sci. 117, 3367–3377 (2004)
Nowak, K. J. et al. Mutations in the skeletal muscle alpha-actin gene in patients with actin myopathy and nemaline myopathy. Nature Genet. 23, 208–212 (1999)
Ohlsson, M., Tajsharghi, H., Darin, N., Kyllerman, M. & Oldfors, A. Follow-up of nemaline myopathy in two patients with novel mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul. Disord. 14, 471–475 (2004)
Ilkovski, B. et al. Evidence for a dominant-negative effect in ACTA1 nemaline myopathy caused by abnormal folding, aggregation and altered polymerization of mutant actin isoforms. Hum. Mol. Genet. 13, 1727–1743 (2004)
Graziano, C., Bertini, E., Minetti, C. & Porfirio, B. Alpha-actin gene mutations and polymorphisms in Italian patients with nemaline myopathy. Int. J. Mol. Med. 13, 805–809 (2004)
Perkins, K. Z. et al. P3.48 Exome sequencing with linkage analysis identifies a novel ACTA1 variant in a large family with progressive muscle weakness. Neuromuscul. Disord. 21, 696–697 (2011)
Nair, U. B. et al. Crystal structures of monomeric actin bound to cytochalasin D. J. Mol. Biol. 384, 848–864 (2008)
Sudo, A. et al. Sibling cases of severe infantile form of nemaline myopathy with ACTA1-gene mutation. [in Japanese]. No To Hattatsu 45, 452–456 (2013)
Yao, X., Grade, S., Wriggers, W. & Rubenstein, P. A. His(73), often methylated, is an important structural determinant for actin. A mutagenic analysis of HIS(73) of yeast actin. J. Biol. Chem. 274, 37443–37449 (1999)
Agrawal, P. B. et al. Heterogeneity of nemaline myopathy cases with skeletal muscle alpha-actin gene mutations. Ann. Neurol. 56, 86–96 (2004)
D'Amico, A. et al. Fatal hypertrophic cardiomyopathy and nemaline myopathy associated with ACTA1 K336E mutation. Neuromuscul. Disord. 16, 548–552 (2006)
Stenzel, W. et al. Fetal akinesia caused by a novel actin filament aggregate myopathy skeletal muscle actin gene (ACTA1) mutation. Neuromuscul. Disord. 20, 531–533 (2010)
Jain, R. K. et al. Nemaline myopathy with stiffness and hypertonia associated with an ACTA1 mutation. Neurology 78, 1100–1103 (2012)
Kollmar, M., Dürrwang, U., Kliche, W., Manstein, D. J. & Kull, F. J. Crystal structure of the motor domain of a class-I myosin. EMBO J. 21, 2517–2525 (2002)
Acknowledgements
We thank O. Hofnagel for excellent assistance in cryo sample preparation and electron microscopy, R. S. Goody for continuous support and for comments on the manuscript and I. Vetter for assistance in data processing. We gratefully acknowledge R. Matadeen and S. de Carlo (FEI Company) for image acquisition at the National Center for Electron Nanoscopy in Leiden (NeCEN), which is co-financed by grants from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (project 175.010.2009.001) and by the European Union’s Regional Development Fund through ‘Kansen voor West’ (project 21Z.014). J.v.d.E. is a fellow of Studienstiftung des deutschen Volkes. This work was supported by the Behrens-Weise foundation (to S.R.), NIH U54 094598 and R01 60635 (to P.A.P.), DFG MA1081/19-1 (to D.J.M.) and R37HL036153 (to W.L.).
Author information
Authors and Affiliations
Contributions
J.v.d.E. and S.R. designed the project. M.M. and D.J.M. provided protein samples. W.L. and D.J.M. provided information on tropomyosin and actin. J.v.d.E. performed research, analysed the data, and prepared figures. J.v.d.E. and P.A.P. improved image processing of helical specimens. S.R. managed the project and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Micrographs and classifications of different data sets.
a–h, Representative digital micrographs and corresponding representative two-dimensional class averages of F-actin decorated with tropomyosin in amorphous ice (in total 300 class averages of randomly chosen 40,000 phase-flipped segments of in total 109,242 segments from 689 images) (a, b), negatively stained (in total 300 class averages of 27,926 segments from 111 images) (c, d), negatively stained after cross-linking with glutardialdehyde (in total 300 class averages of 27,011 segments from 81 images) (e, f), and a micrograph of negatively stained bare F-actin (in total 100 class averages of 8,371 segments from 40 images) (g, h). Scale bars, 50 nm. Each class average (‘Classifications’) contains 130–200 (cryo data set) or 70–90 (negatively stained data sets) single segments. Scale bars, 10 nm. A boxed region of the digital micrograph in a was band-pass filtered to allow a better visualization of the filaments. Insets in c, e and g show 2× magnified regions of the digital micrographs. i, Two of the class averages depicted in b that show a clear tropomyosin density. The tropomyosin density is indicated by black arrow heads. Scale bar, 10 nm. j, Comparison of representative class averages of the three negative-stain data sets. Class averages of the data sets with tropomyosin (top panels) show additional density and a larger diameter than bare F-actin (bottom panel). Scale bar, 10 nm.
Extended Data Figure 2 Resolution of the F-actin–tropomyosin complex.
a, FSC curves of different areas of interest (green) by masking (see Methods). The resolution of tropomyosin was estimated using the twice down-sampled data set (2.24 Å per pixel). The FSC0.5 criterion indicates that the tropomyosin density map has a resolution of 6.5 Å. The resolution of the final F-actin density map is estimated at a resolution of 3.7 Å. b, Surfaces of F-actin with B-factors (high is red, low is blue) estimated by the reciprocal space refinement in PHENIX42. A side view as well as top views on the inward- and outward-facing surfaces, that is, facing the adjacent F-actin subunit inside the filament and oriented to the periphery, respectively, are shown. c, d, Putative structures of the C and N termini (cyan), respectively. Map density is missing in these regions.
Extended Data Figure 3 Representative regions of the F-actin cryo-EM map.
a, Overview of the atomic model of an F-actin subunit rainbow-coloured from the N terminus (blue) to the C terminus (red). ADP and the coordinated cation, probably Mg2+, are depicted inside their corresponding map densities. b–d, Side views. Interface between the D-loop (SD2) and the SD3 of the adjacent actin subunit (b), outer two helices of SD1 (c), and inner β-sheet with N terminus (d). e, f, Front views. Connection of SD1 and SD3 (e) and nucleotide binding cleft (f). g, h, Side views. Inner β-sheet of SD3 with one highlighted strand (g) and outer part of SD3 (h). i, j, Back views. SD4 separated in two parts.
Extended Data Figure 4 Overview of inter- and intrastrand interactions and hydrophobic D-loop.
a, Table of identified residues that are involved in intra- and intermolecular interactions and known mutations. Mutation data are from refs 3, 15, 20 and 52, 53, 54, 55, 56, 57, 58. b, Overview of the D-loop bound to the hydrophobic cleft in SD3 of the F-actin molecule on top. Surfaces and residues are coloured from high (yellow) to low (white) hydrophobicity. c, Involved hydrophobic residues of the D-loop. d, e, Modifications in the D-loop, like oxidation of methionine 44 (ref. 20) (d) or mutations of methionine 44 and glycine 46 (M44T, G46G)15,20 (e) change the polarity or insert charges and consequently weaken the hydrophobic interactions. Thus, this destabilization of the intrastrand contact is connected to anomalous actin filament assembly and nemaline myopathies.
Extended Data Figure 5 Inter- and intrastrand F-actin interactions.
a–e, The interface at the plug involves three residues (R39, E270, D286) that form salt bridges and mediate not only one interstrand contact but also one intrastrand contact (see also Fig. 2a). In addition, the orientation of residues 264–269 result in a negatively charged patch that electrostatically interacts with positively charged residues on the opposing actin. a, b, Front and back view of the interface at the plug, respectively. c–e, Surface representations (front views in c and d, back view in e) depicting the Coulomb potential (ranging from −10 kcal mol−1 (red) to +10 kcal mol−1 (blue) at pH 7.5), indicating that the interaction of the upper region of the plug with adjacent inter- or intrastrand molecules is mediated by electrostatic interactions. f, Another interstrand contact is formed by residues 110–115 of SD1 and residues 191–199 of SD4 of the adjacent actin. However, no prominent electrostatic or hydrophobic interactions could be identified at this interface.
Extended Data Figure 6 Nucleotide binding site and intramolecular interactions.
a, Table of identified intramolecular interactions that result in a stabilization of the nucleotide binding cleft, coordination of ADP and a divalent cation. Mutation data are from refs 3, 15, 23, 53, 54, 56, 57 and 60, 61, 62, 63, 64. b, Coordination of ADP and Ca2+ in the nucleotide binding cleft in G-actin (PDB accession code 3EL2; ref. 59). c–e, Back views of the nucleotide binding cleft of F-actin (cyan) with bound ADP–Mg2+ or ADP–Ca2+ (c), ATP–Ca2+ (d, relative position taken from PDB accession code 3EL2; ref. 59) and conformational changes between the G-actin-ATP and F-actin-ADP state (e). Glutamine 137 is moved closer to ADP, coordinating not only the cation (as in G-actin) but also the nucleotide β-phosphate (c). The presence of ATP instead of ADP in the nucleotide-binding site would be sterically unfavourable, suggesting that a different intermediate conformation exists for F-actin-ATP (d). The shorter distance of glutamine 137 to the γ-phosphate probably induces ATP hydrolysis and then afterwards the cation takes the position of the γ-phosphate in the ADP-state (e). For comparison the position of glutamine 137 in G-actin is shown in yellow and the transition from G-actin to F-actin is depicted by arrows. f, Mutation of glutamine 137 to histidine results in hampered coordination of the ion and the nucleotide and is connected to nemaline myopathies23.
Extended Data Figure 7 Model of barbed-end and pointed-end binding and G- to F-actin transition based on a comparison of start point (G-actin) and end point (F-actin).
a, b, Binding of new G-actin (yellow, PDB accession code 3EL2; ref. 59) at the barbed end of F-actin (green) is initiated by the intrastrand binding of SD4 of G-actin to SD3 of F-actin (a, side view) and the interstrand binding of SD4 and SD1 (b, front view), respectively. G-actin is overlaid with a subunit of the structure found in F-actin (cyan). The main interstrand contacts are already present at the start of the transition and thereby guide the binding G-actin to its correct position, determining the symmetry of the filament. c, After initial binding the D-loop is trapped in the hydrophobic cleft of SD3 of F-actin and pulls on SD2. F-actin is depicted in surface representation (green). d, Finally, the SD2 and concomitantly SD1 are rotated and the final F-actin conformation is stabilized by various intramolecular interactions (see Figs 2c–f and 3c–f, Extended Data Fig. 6a). e, Owing to the relatively large distance from the SD1 and SD2 subdomains of the newly bound G-actin to the subunit of the opposite strand, the only influence of the opposite strand on the binding of G-actin is at the docking position of the D-loop. The transition of R39 illustrates well the docking of the D-loop. The residue is depicted in both states (G-actin and F-actin). f–h, Binding of new G-actin (yellow, PDB accession code 3EL2; ref. 59) at the pointed end of F-actin (green) is initiated by initial intrastrand binding (g) of SD3 of G-actin to SD4 of F-actin at the pointed end and interstrand binding of SD4 to SD3 (h), respectively. Again, the main interstrand contacts are available before the transition of G-actin (yellow) to F-actin (SD1 and SD2 in cyan, SD3 and SD4 in red). i, During binding to F-actin the transition from G-actin to F-actin is initiated by an induced fit of the F-actin D-loop to the hydrophobic cleft of the newly bound G-actin. This leads to a pulling down of the central β-sheet of SD3 of G-actin. j, The β-sheet is thereby straightened and pushes up two adjacent helices of SD1. k, The slight dislocation of these helices is transmitted to other regions of SD1 and thereby emphasized. This leads to a global rotation of SD1, which results in a considerable rotation of SD2 by an angle of 20° and a closure of the nucleotide binding cleft (Fig. 3; see Supplementary Video 3).
Extended Data Figure 8 Tropomyosin binding and comparison of reconstructions regarding the tropomyosin position on F-actin.
a, Table showing putative residues of F-actin involved in tropomyosin binding and known mutations. Mutation data are from refs 15 and 65. b–e, Reconstructions of F-actin decorated with tropomyosin calculated from: the cryo-EM data set filtered to 15 Å (b), from a negatively stained data set (c), from a negatively stained data set after cross-linking with glutardialdehyde (d), and from negatively stained bare F-actin (e). f, By calculating a difference map between tropomyosin–bare F-actin (blue) and glutardialdehyde–bare F-actin (green), differences in the tropomyosin position on bare F-actin (grey) are visualized. g, Overlay of difference maps showing that the position of tropomyosin in the cryo-EM reconstruction (yellow) corresponds to the tropomyosin position of the negatively stained data set with the cross-linked complex (green). h, Surface of F-actin and tropomyosin (pseudo-repeats 2–6) with the electrostatic Coulomb potentials at pH 7.5 and pH 4, ranging from −10 kcal mol−1 (red) to +10 kcal mol−1 (blue) (see also Fig. 4b). Tropomyosin was rotated by 180° and shifted to the right to allow a better view of the F-actin–tropomyosin interface. Difference maps of the glutardialdehyde–bare F-actin and the tropomyosin–bare F-actin map are shown on the F-actin surface at pH 7.5 (left) and pH 4.0 (middle), respectively.
Extended Data Figure 9 Model of tropomyosin transition on F-actin during myosin binding.
a, Cryo-EM structure of the F-actin–tropomyosin complex with tropomyosin in the A-state. Tropomyosin (yellow, A-state), F-actin (green). b–d, Initial weak binding of myosin (magenta, PDB accession code 1LKX; ref. 66) to the F-actin–tropomyosin filament in the absence of troponin. Most of the myosin binding sites on F-actin are not occupied by tropomyosin and only loop 4 and the cardiomyopathy loop are sterically hindered from binding to the F-actin filament (b). Actin-induced closure of the 50-kDa cleft of myosin (c) results in a strong binding of myosin and tropomyosin moves to its M-state position (blue) (d). e–g, F-actin–tropomyosin–myosin complex in the rigor state (PDB accession code 4A7H; ref. 8). Myosin is shown in red. h–l, There are two possible ways for the transition of tropomyosin from the A-state to the M-state. Tropomyosin either rolls (i, j) or slides (k, l) from one to the other position. Rolling would involve an azimuthal rotation of ∼16° with respect to the F-actin axis (inset of j) and a left-handed rotation of ∼70° (indicated by solid arrows) or a right-handed rotation of ∼110° (indicated by dotted arrows) with respect to its own axis (i, j). Sliding would imply not only an azimuthal shift of ∼12 Å (indicated by white arrows in k), but also a tremendous shift of a half-tropomyosin repeat (that is, ∼35 Å) along the F-actin filament (k, l). The radius to the filament axis would be preserved in both situations. The inset of l depicts the vectors for a shifting transition of tropomyosin: an azimuthal and longitudinal shift of 12 Å and 35 Å, resulting in an overall shift of 37 Å.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion and an additional reference. (PDF 179 kb)
Cryo-EM structure of F-actin decorated with tropomyosin
a: Cryo-EM density of F-actin (grey, five central subunits: green, central subunit: cyan) in complex with tropomyosin (yellow) at an average resolution of 3.7 Å for F-actin and 6.5 Å for tropomyosin b: Zoom on the central five F-actin subunits. c: Zoom on the central F-actin subunit. (MOV 27110 kb)
Atomic model of F-actin in the cryo-EM density map
a: Cryo-EM density of an F-actin subunit with the respective atomic model. b: Zoom on α-helices of the nucleotide binding cleft with ADP and the coordinated cation, probably Mg2+. c: Zoom on the β sheet of SD3. d: Atomic model of an F-actin subunit rainbow coloured from the N-terminus (blue) to the C-terminus (red). (MOV 26400 kb)
Conformational changes between G-actin-ATP and F-actin-ADP
a-c: Morph between G-actin (yellow, PDB: 1J6Z) to F-actin (cyan) shown in side, top and front view, respectively. d: Zoom on the nucleotide binding cleft highlighting Q137, which has been shown to play a central role in ATP hydrolysis. For better illustration, the F-actin model has been hidden. After the transition to F-actin, ATP and the coordinated cation are cross-faded to ADP and the coordinated cation. (MOV 27281 kb)
Model of tropomyosin transition on F-actin during myosin binding
a: Cryo-EM structure of the F-actin-tropomyosin complex with tropomyosin in the A-state (yellow) and M-state (PDB: 4A7H) (blue), F-actin (green). b-d: Side and bottom views on myosin (magenta, PDB: 1LKX) binding to the F-actin-tropomyosin filament in the absence of troponin. Actin-induced closure of the 50-kDa cleft of myosin results in a strong binding of myosin and tropomyosin moves to its M-state position (blue). There are two possible ways for the transition of tropomyosin between the two states. The video shows only the right-handed rotation of ∼ 110°. (MOV 26987 kb)
Rights and permissions
About this article
Cite this article
von der Ecken, J., Müller, M., Lehman, W. et al. Structure of the F-actin–tropomyosin complex. Nature 519, 114–117 (2015). https://doi.org/10.1038/nature14033
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14033
This article is cited by
-
LAMB3 Promotes Myofibrogenesis and Cytoskeletal Reorganization in Endometrial Stromal Cells via the RhoA/ROCK1/MYL9 Pathway
Cell Biochemistry and Biophysics (2024)
-
Actin cytoskeleton and complex cell architecture in an Asgard archaeon
Nature (2023)
-
50 Years of the steric-blocking mechanism in vertebrate skeletal muscle: a retrospective
Journal of Muscle Research and Cell Motility (2023)
-
Quantitative proteomics and in-cell cross-linking reveal cellular reorganisation during early neuronal differentiation of SH-SY5Y cells
Communications Biology (2022)
-
The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.