Abstract
T-helper type 17 (TH17) cells that produce the cytokines interleukin-17A (IL-17A) and IL-17F are implicated in the pathogenesis of several autoimmune diseases1,2. The differentiation of TH17 cells is regulated by transcription factors such as RORγt3,4, but post-translational mechanisms preventing the rampant production of pro-inflammatory IL-17A have received less attention. Here we show that the deubiquitylating enzyme DUBA is a negative regulator of IL-17A production in T cells. Mice with DUBA-deficient T cells developed exacerbated inflammation in the small intestine after challenge with anti-CD3 antibodies. DUBA interacted with the ubiquitin ligase UBR5, which suppressed DUBA abundance in naive T cells. DUBA accumulated in activated T cells and stabilized UBR5, which then ubiquitylated RORγt in response to TGF-β signalling. Our data identify DUBA as a cell-intrinsic suppressor of IL-17 production.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hemdan, N. Y. A. et al. Interleukin-17-producing T helper cells in autoimmunity. Autoimmun. Rev. 9, 785–792 (2010)
Miossec, P. & Kolls, J. K. Targeting IL-17 and TH17 cells in chronic inflammation. Nature Rev. Drug Discov. 11, 763–776 (2012)
Yosef, N. et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468 (2013)
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012)
Komander, D., Clague, M. J. & Urbé, S. Breaking the chains: structure and function of the deubiquitinases. Nature Rev. Mol. Cell Biol. 10, 550–563 (2009)
Kayagaki, N. et al. DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 (2007)
Huang, O. W. et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nature Struct. Mol. Biol. 19, 171–175 (2012)
Lee, P. P. et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001)
Liang, S. C. et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 (2006)
Esplugues, E. et al. Control of TH17 cells occurs in the small intestine. Nature 475, 514–518 (2011)
Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30, 647–675 (2012)
Georgiades, P. et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 34, 251–256 (2002)
Chabaud, M., Fossiez, F., Taupin, J. L. & Miossec, P. Enhancing effect of IL-17 on IL-1-induced IL-6 and leukemia inhibitory factor production by rheumatoid arthritis synoviocytes and its regulation by Th2 cytokines. J. Immunol. 161, 409–414 (1998)
Chabaud, M. et al. Human interleukin-17: A T cell-derived proinflammatory cytokine produced by the rheumatoid synovium. Arthritis Rheum. 42, 963–970 (1999)
Lock, C. et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nature Med. 8, 500–508 (2002)
Matusevicius, D. et al. Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Mult. Scler. 5, 101–104 (1999)
Yang, X. O. et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008)
Voo, K. S. et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc. Natl Acad. Sci. USA 106, 4793–4798 (2009)
Kryczek, I. et al. IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J. Immunol. 186, 4388–4395 (2011)
Beriou, G. et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood 113, 4240–4249 (2009)
Ma, C. & Dong, X. Colorectal cancer-derived Foxp3+ IL-17+ T cells suppress tumour-specific CD8+ T cells. Scand. J. Immunol. 74, 47–51 (2011)
Powrie, F., Leach, M. W., Mauze, S., Caddle, L. B. & Coffman, R. L. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 5, 1461–1471 (1993)
Mottet, C., Uhlig, H. H. & Powrie, F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J. Immunol. 170, 3939–3943 (2003)
Ghoreschi, K. et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010)
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011)
Zhang, P. et al. PARP-1 regulates expression of TGF-β receptors in T cells. Blood 122, 2224–2232 (2013)
Sterling, J. A., Wu, L. & Banerji, S. S. PARP regulates TGF-β receptor type II expression in estrogen receptor-positive breast cancer cell lines. Anticancer Res. 26, 1893–1901 (2006)
Lönn, P. et al. PARP-1 attenuates Smad-mediated transcription. Mol. Cell 40, 521–532 (2010)
Callaghan, M. J. et al. Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17, 3479–3491 (1998)
siRNA. stabilization prolongs gene knockdown in primary T lymphocytes. Eur. J. Immunol. 38, 2616–2625 (2008)
Collison, L. W. & Vignali, D. A. A. In vitro Treg suppression assays. Methods Mol. Biol. 707, 21–37 (2011)
Zheng, Y. et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007)
Anania, V. G. et al. Peptide level immunoaffinity enrichment enhances ubiquitination site identification on individual proteins. Mol. Cell. Proteomics 13, 145–156 (2014)
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003)
Beausoleil, S. A., Villén, J., Gerber, S. A., Rush, J. & Gygi, S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature Biotechnol. 24, 1285–1292 (2006)
Bakalarski, C. E. et al. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J. Proteome Res. 7, 4756–4765 (2008)
Kirkpatrick, D. S. et al. Phosphoproteomic characterization of DNA damage response in melanoma cells following MEK/PI3K dual inhibition. Proc. Natl Acad. Sci. USA 110, 19426–19431 (2013)
Acknowledgements
We thank A. C. Chan for discussions, Q. Song for statistical analysis, and C. Bakalarski for informatics support.
Author information
Authors and Affiliations
Contributions
S.R., N.K., Q.T.P., C.E., R.N., X.W., J.L., R.L., O.W.H., J.D. and Z.M. performed experiments designed and analysed by S.R., D.S.K., J.R.L., A.G.C., W.O. and V.M.D.; J.W. and L.D. performed histopathology; M.V. synthesized siRNAs; S.R., W.O. and K.N. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
All authors were employees of Genentech, Inc.
Extended data figures and tables
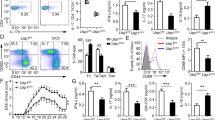
Extended Data Figure 1 Analysis of the T-cell compartment in Dubafl/fl CD4-Cre mice aged 6 weeks.
a, Targeting strategy for the Dubafl allele. The PGK-neo selection cassette was removed with an Flp deleter strain. b, Immunoblots of immune cell subsets from Duba+/+ CD4-Cre (wild-type, WT) and Dubafl/fl CD4-Cre (knockout, KO) mice. CD11c+ dendritic cells (DC) and peritoneal macrophages (MF) were stimulated with 50 ng ml−1 lipopolysaccharide (LPS) overnight. c–j, Knockout and wild-type mice were aged 6 weeks. n = 5 mice per genotype. Leukocyte numbers in the thymus, spleen and mesenteric lymph node (c), CD4−CD8− (DN) and CD4+CD8+ (DP) cells in the thymus enumerated by cell counting and flow cytometry (d), Numbers of CD4+CD8− and CD4−CD8+ cells in thymus (e), in mesenteric lymph nodes (mLN) (f) and spleen (g). h, Percentages of naive (CD45RbhiCD44low) and memory (CD45RblowCD44hi) CD4+ T cells. i, FOXP3+ T cells in the thymus. j, Percentage of FOXP3+ T cells in the spleen and mesenteric lymph nodes. Each circle represents one mouse. Error bars, s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
Extended Data Figure 2 Increased IL-17A production by CD4+ and CD8+ T cells from Dubafl/fl CD4-Cre mice.
a, b, Percentage of splenic CD4+ T cells from wild-type (Duba+/+ CD4-Cre) or knockout (Dubafl/fl CD4-Cre) mice that stained positive for intracellular IFN-γ, IL-4 or IL-17A (a) or RORγt (b) by flow cytometry. c, d, Percentage of CD4+ T cells expressing IFN-γ, IL-4 and IL-17A (c) or RORγt (d) after collection on day 8 after immunization with OVA/CFA, and subsequent culture with PMA and ionomycin for 4 h. e, Percentage of splenic CD8+ T cells from knockout and wild-type mice that stained positive for intracellular IL-17A by flow cytometry. Representative contour plots are shown. Each circle represents one mouse. Error bars, s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test).
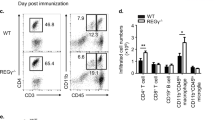
Extended Data Figure 3 Cytokine production by Duba+/+ CD4-Cre and Dubafl/fl CD4-Cre mice injected with anti-CD3 antibodies.
a–c, CD4+ T cells expressing IL-17A (a), RORγt (b) or IFN-γ (c) by intracellular staining and flow cytometry. Cells from the spleen or mesenteric lymph node were isolated from wild-type (Duba+/+ CD4-Cre) or knockout (Dubafl/fl CD4-Cre) mice at 48 h after injection with anti-CD3 antibodies and then stimulated with PMA and ionomycin for 4 h. Representative contour plots are shown. d, Serum cytokines at 48 h after injection of anti-CD3 antibodies. PBS indicates wild-type mice injected with vehicle alone. nd, not detected. e, Intraepithelial lymphocytes from the small intestine producing IFN-γ based on intracellular staining and flow cytometry. Cells were collected at 48 h after injection and stimulated with PMA and ionomycin for 4 h. f, Small intestine weights at 48 h after injection of anti-CD3 antibodies. Each circle represents one mouse. Error bars, s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test). Data are representative of 2 (a–d, f) or 3 (e) independent experiments.
Extended Data Figure 4 DUBA-deficient Treg cells express RORγt and produce IL-17A, but are suppressive in vitro and in vivo.
a, b, Percentage of RORγt+ (a) or IL-17A-producing (b) Treg cells in the spleen or mesenteric lymph node of naive mice. c, IL-17A secretion by CD25+ Treg cells after stimulation with anti-CD3 and anti-CD28 antibodies for 24 h. Data are representative of two independent experiments. d, Proliferation of C57BL/6 CD4+ T cells (responder T cells; Tresp) co-cultured with Treg cells in the ratios indicated. Division index denotes the average number of cell divisions that a cell in the original population has undergone. Data are representative of two independent experiments. e, Weight relative to time zero of Rag2−/− mice after transfer of either naive CD4+ T cells alone or together with Duba+/+ or Duba−/− Treg cells. f, Animal clinical scores. g, Colon weights at 10 weeks after T-cell transfer. h, Colon histology scores at 10 weeks. n = 10 (cell transfer groups), n = 5 (PBS). Error bars, s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t-test). Data are representative of two independent experiments.
Extended Data Figure 5 Composition of the T-cell compartment in Dubafl/fl CD4-Cre mice aged one year.
a, CD4+ and CD8+ T cells enumerated by cell counting and flow cytometry. b–f, Percentages of FOXP3+ Treg cells (b), RORγt+ T cells (c), RORγt+ Treg cells (d), naive (CD45RbhiCD44low) CD4+ T cells (e), and memory (CD45RblowCD44hi) CD4+ T cells (f) in the spleen and mesenteric lymph node. g, Percentages of IFN-γ, IL-4 or IL-17-producing CD4+ T cells in spleen. Each circle represents one mouse. Error bars, s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (unpaired Student’s t-test).
Extended Data Figure 6 Cytokine production and lineage transcription factor expression in DUBA-deficient T cells.
a, Immunoblots of T-helper subsets after 24 h of culture. Graphs indicate relative DUBA abundance from densitometry measurements. Error bars, s.e.m. of three independent experiments. b, Representative contour plots indicating the percentage of IL-17A-expressing TH17 and iTreg cells. c, Expression of Il17f, Il21, Il9, Il22 and Il10 mRNAs by Duba+/+ and Duba−/− T cells that were stimulated with anti-CD3 and anti-CD28 antibodies in the presence of TGF-β and IL-6. Secreted cytokines were measured on the third day of culture. d, IL-17A secretion by Duba+/+ or Duba−/− naive CD4+ T cells after culture with TGF-β and IL-6 for 6 days and then secondary stimulation with anti-CD3 and anti-CD28 antibodies for 2 days. After a 6-day secondary stimulation, the cells were re-stimulated again. e Percentage of CD8+ T cells expressing IL-17A after 5 days of culture under Tc0, Tc1, Tc2 and Tc17 polarizing conditions by flow cytometry. Representative contour plots under Tc17 conditions are shown. f, Flow cytometric analysis of T-bet, GATA3, RORγt and FOXP3 expression in TH11, TH2 and iTreg cells. Error bars, s.e.m. of triplicate measurements. Data are representative of two independent experiments.
Extended Data Figure 7 Normal expression of TH17 factors in DUBA-deficient TH17 cells.
a–c, Expression of TH17 factors by Duba+/+ and Duba−/− CD4+ T cells that were stimulated with anti-CD3 and anti-CD28 antibodies in the presence of TGF-β and IL-6. Graphs show mRNA expression (a) and immunoblots indicate protein expression (b), immunoblots indication expression of retinoid acid receptors (c). Data are representative of two independent experiments.
Extended Data Figure 8 Normal TGF-β receptor expression and SMAD activation in DUBA-deficient TH17 cells.
a, Expression of TGF-βRI and TGF-βRII in Duba+/+ and Duba−/− TH17 cells after culture for the times indicated. Graphs indicate mRNA expression, whereas immunoblots indicate protein expression. Error bars, s.e.m. of duplicate measurements. Data are representative of two independent experiments. b, Immunoblots of the cells in a. c, Immunoblots of Duba+/+ or Duba−/− TH17 cells after stimulation with TGF-β and IL-6.
Extended Data Figure 9 Identification of DUBA substrates in TH17 cells by K-ε-GG peptide analysis.
a, Volcano plots of ubiquitinated proteins identified after immunoprecipitating K-ε-GG peptides from Duba+/+ and Duba−/− TH17 cells after 4 days of culture. P values are plotted against fold-change in number of peptides for each protein. b, Proteins >1.5-fold differentially ubiquitinated in Duba+/+ and Duba−/− TH17 cells (green and red denote increased and decreased ubiquitylation in the knockout, respectively).
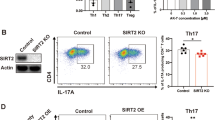
Extended Data Figure 10 The roles of PARP-1 and UBR5 in the regulation of Il-17A production in T cells.
a, Mass spectrometry detection of PARP-1 co-immunoprecipitated with Flag-tagged wild-type DUBA or a DUBA(Cys224Ser) mutant expressed in TH17 cells. Graph indicates the number of unique peptides in experimental samples (black) or control cells expressing GFP (white). b, IL-17A production by TH17 cells or iTreg cells (TGF-β plus IL-2) after 3 days of culture in DMSO vehicle or veliparib. c, Expression of Il17a and Rorc mRNA in TH17 cells treated with veliparib or DMSO vehicle. d, Immunoblots of RORγt in TH17 cells cultured for 2 days with either DMSO or veliparib, and then treated for the indicated times in cycloheximide. Graphs indicate relative RORγt abundance from densitometry measurements. Error bars, s.e.m. of three independent experiments. e, Immunoblot of Duba+/+ and Duba−/− TH17 cells cultured for the times indicated. Error bars, s.e.m. of triplicate measurements. **P < 0.01 (unpaired Student’s t-test). f, Immunoblot of UBR5 co-immunoprecipitated with Flag-tagged DUBA from retrovirally transduced CD4+ T cells. Control cells were transduced with GFP-expressing virus. g, Immunoblot of UBR5 in wild-type TH17 cells transfected with the siRNA indicated. h, IL-17A production by wild-type TH17 cells transfected with the siRNA indicated. Error bars, s.e.m. of two independent experiments. i, Expression of Ubr5 mRNA in CD4+ T cells. Error bars, s.e.m of three independent experiments. j, Immunoblots of wild-type T cells stimulated with anti-CD3 or anti-CD28 antibodies for the times indicated.
Rights and permissions
About this article
Cite this article
Rutz, S., Kayagaki, N., Phung, Q. et al. Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421 (2015). https://doi.org/10.1038/nature13979
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13979
This article is cited by
-
UBR5 promotes antiviral immunity by disengaging the transcriptional brake on RIG-I like receptors
Nature Communications (2024)
-
Thiostrepton alleviates experimental colitis by promoting RORγt ubiquitination and modulating dysbiosis
Cellular & Molecular Immunology (2023)
-
Autophagy of OTUD5 destabilizes GPX4 to confer ferroptosis-dependent kidney injury
Nature Communications (2023)
-
DHX9/DNA-tandem repeat-dependent downregulation of ciRNA-Fmn1 in the dorsal horn is required for neuropathic pain
Acta Pharmacologica Sinica (2023)
-
The DUBA-SLC7A11-c-Myc axis is critical for stemness and ferroptosis
Oncogene (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.